Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
| Table of Contents | ||
|---|---|---|
|
| Anchor | ||||
|---|---|---|---|---|
|

Figure 1. Delta System
Delta is supported by the National Science Foundation under Grant No. OAC-2005572.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
| Delta is now accepting proposals. |
|---|
Status Updates and Notices
Delta is tentatively scheduled to enter production in Q3 2022.
| Info | ||
|---|---|---|
| ||
| Items in light grey font are in progress and coming soon. Example software or feature not yet implemented. |
Introduction
Delta is a dedicated, ACCESS-allocated resource allocated resource designed by HPE and NCSA, delivering a highly capable GPU-focused compute environment for GPU and CPU workloads. Besides offering a mix of standard and reduced precision GPU resources, Delta also offers GPU-dense nodes with both NVIDIA and AMD GPUs. Delta provides high performance node-local SSD scratch filesystems, as well as both standard Lustre and relaxed-POSIX parallel filesystems spanning the entire resource.
Delta's CPU nodes are each powered by two 64-core AMD EPYC 7763 ("Milan") processors, with 256 GB of DDR4 memory. The Delta GPU resource has four node types: one with 4 NVIDIA A100 GPUs (40 GB HBM2 RAM each) connected via NVLINK and 1 64-core AMD EPYC 7763 ("Milan") processor, the second with 4 NVIDIA A40 GPUs (48 GB GDDR6 RAM) connected via PCIe 4.0 and 1 64-core AMD EPYC 7763 ("Milan") processor, the third with 8 NVIDIA A100 GPUs in a dual socket AMD EPYC 7763 (128-cores per node) node with 2 TB of DDR4 RAM and NVLINK, and the fourth with 8 AMD MI100 GPUs (32GB HBM2 RAM each) in a dual socket AMD EPYC 7763 (128-cores per node) node with 2 TB of DDR4 RAM and PCIe 4.0.
Delta has 124 standard CPU nodes, 100 4-way A100-based GPU nodes, 100 4-way A40-based GPU nodes, 5 8-way A100-based GPU nodes, and 1 8-way MI100-based GPU node. Every Delta node has high-performance node-local SSD storage (740 GB for CPU nodes, 1.5 TB for GPU nodes), and is connected to the 7 PB Lustre parallel filesystem via the high-speed interconnect. The Delta resource uses the SLURM workload manager for job scheduling.
Delta supports the ACCESS core software stack, including remote login, remote computation, data movement, science workflow support, and science gateway support toolkits.
Account Administration
- For ACCESS projects please use the ACCESS user portal for project and account management.
- Non-ACCESS Account and Project administration, such as adding a someone to a project, is handled by NCSA Identity and NCSA group management tools. For more information, please see the NCSA Allocation and Account Management documentation page.
Configuring Your Account
- Bash is the default shell, submit a support request to change your default shell
- Environment variables: ACCESS CUE, SLURM batch
- Using Modules
System Architecture
Delta is designed to help applications transition from CPU-only to GPU or hybrid CPU-GPU codes. Delta has some important architectural features to facilitate new discovery and insight:
- A single processor architecture (AMD) across all node types: CPU and GPU
- Support for NVIDIA A100 MIG GPU partitioning allowing for fractional use of the A100s if your workload isn't able to exploit an entire A100 efficiently
- Ray tracing hardware support from the NVIDIA A40 GPUs
- Nine large memory (2 TB) nodes
- A low latency and high bandwidth HPE/Cray Slingshot interconnect between compute nodes
- Lustre for home, projects and scratch file systems
- support for relaxed and non-posix IO
- Shared-node jobs and the single core and single MIG GPU slice
- Resources for persistent services in support of Gateways, Open OnDemand, and Data Transport nodes.
- Unique AMD MI-100 resource
Model Compute Nodes
The Delta compute ecosystem is composed of five node types:
- Dual-socket CPU-only compute nodes
- Single socket 4-way NVIDIA A100 GPU compute nodes
- Single socket 4-way NVIDIA A40 GPU compute nodes
- Dual-socket 8-way NVIDIA A100 GPU compute nodes
- Single socket 8-way AMD MI100 GPU compute nodes
The CPU-only and 4-way GPU nodes have 256 GB of RAM per node while the 8-way GPU nodes have 2 TB of RAM. The CPU-only node has 0.74 TB of local storage while all GPU nodes have 1.5 TB of local storage.
Table. CPU Compute Node Specifications
| Specification | Value |
|---|---|
Number of nodes | 132 |
| CPU | AMD EPYC 7763 "Milan" (PCIe Gen4) |
| Sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 0.74 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 4-way NVIDIA A40 GPU Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 100 |
| GPU | NVIDIA A40 |
| GPUs per node | 4 |
| GPU Memory (GB) | 48 DDR6 with ECC |
| CPU | AMD Milan |
| CPU sockets per node | 1 |
Cores per socket | 64 |
| Cores per node | 64 |
Hardware threads per core | 1 |
Hardware threads per node | 64 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
The A40 GPUs are connected via PCIe Gen4 and have the following affinitization to NUMA nodes on the CPU. Note that the relationship between GPU index and NUMA domain are inverse.
Table. 4-way NVIDIA A40 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | SYS | SYS | SYS | SYS | 48-63 | 3 |
| GPU1 | SYS | X | SYS | SYS | SYS | 32-47 | 2 |
| GPU2 | SYS | SYS | X | SYS | SYS | 16-31 | 1 |
| GPU3 | SYS | SYS | SYS | X | PHB | 0-15 | 0 |
| HSN | SYS | SYS | SYS | PHB | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 4-way NVIDIA A100 GPU Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 100 |
| GPU | NVIDIA A100 |
| GPUs per node | 4 |
| GPU Memory (GB) | 40 |
| CPU | AMD Milan |
| CPU sockets per node | 1 |
Cores per socket | 64 |
| Cores per node | 64 |
Hardware threads per core | 1 |
Hardware threads per node | 64 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 4-way NVIDIA A100 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | NV4 | NV4 | NV4 | SYS | 48-63 | 3 |
| GPU1 | NV4 | X | NV4 | NV4 | SYS | 32-47 | 2 |
| GPU2 | NV4 | NV4 | X | NV4 | SYS | 16-31 | 1 |
| GPU3 | NV4 | NV4 | NV4 | X | PHB | 0-15 | 0 |
| HSN | SYS | SYS | SYS | PHB | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 8-way NVIDIA A100 GPU Large Memory Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 6 |
| GPU | NVIDIA A100 |
| GPUs per node | 8 |
| GPU Memory (GB) | 40 |
| CPU | AMD Milan |
| CPU sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 2,048 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 8-way NVIDIA A100 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | GPU4 | GPU5 | GPU6 | GPU7 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 48-63 | 3 |
| GPU1 | NV12 | X | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 48-63 | 3 |
| GPU2 | NV12 | NV12 | X | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 16-31 | 1 |
| GPU3 | NV12 | NV12 | NV12 | X | NV12 | NV12 | NV12 | NV12 | SYS | 16-31 | 1 |
| GPU0 | NV12 | NV12 | NV12 | NV12 | X | NV12 | NV12 | NV12 | SYS | 112-127 | 7 |
| GPU1 | NV12 | NV12 | NV12 | NV12 | NV12 | X | NV12 | NV12 | SYS | 112-127 | 7 |
| GPU2 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | X | NV12 | SYS | 80-95 | 5 |
| GPU3 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | X | SYS | 80-95 | 5 |
| HSN | SYS | SYS | SYS | SYS | SYS | SYS | SYS | SYS | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 8-way AMD MI100 GPU Large Memory Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 1 |
| GPU | AMD MI100 |
| GPUs per node | 8 |
| GPU Memory (GB) | 32 |
| CPU | AMD Milan |
| CPU sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 2,048 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
Login Nodes
Login nodes provide interactive support for code compilation.
Specialized Nodes
Delta will support data transfer nodes (serving the "NCSA Delta" Globus Online collection) and nodes in support of other services.
Network
Delta is connected to the NPCF core router & exit infrastructure via two 100Gbps connections, NCSA's 400Gbps+ of WAN connectivity carry traffic to/from users on an optimal peering.
Delta resources are inter-connected with HPE/Cray's 100Gbps/200Gbps SlingShot interconnect.
File Systems
Note: Users of Delta have access to 3 file systems at the time of system launch, a fourth relaxed-POSIX file system will be made available at a later date.
Delta
The Delta storage infrastructure provides users with their $HOME and $SCRATCH areas. These file systems are mounted across all Delta nodes and are accessible on the Delta DTN Endpoints. The aggregate performance of this subsystem is 70GB/s and it has 6PB of usable space. These file systems run Lustre via DDN's ExaScaler 6 stack (Lustre 2.14 based).
Hardware:
DDN SFA7990XE (Quantity: 3), each unit contains
- One additional SS9012 enclosure
- 168 x 16TB SAS Drives
- 7 x 1.92TB SAS SSDs
The $HOME file system has 4 OSTs and is set with a default stripe size of 1.
The $SCRATCH file system has 8 OSTs and has Lustre Progressive File Layout (PFL) enabled which automatically restripes a file as the file grows. The thresholds for PFL striping for $SCRATCH are
| File size | stripe count |
|---|---|
| 0-32M | 1 OST |
| 32M-512M | 4 OST |
| 512M+ | 8 OST |
Best Practices
- To reduce the load on the file system metadata services, the ls option for context dependent font coloring, --color, is disabled by default.
Future Hardware:
An additional pool of NVME flash from DDN has been installed in early summer 2022. This flash is initially deployed as a tier for "hot" data in scratch. This subsystem will have an aggregate performance of 500GB/s and will have 3PB of raw capacity. As noted above this subsystem will transition to an independent relaxed POSIX namespace file system, communications on that timeline will be announced as updates are available.
Taiga
Taiga is NCSA’s global file system which provides users with their $WORK area. This file system is mounted across all Delta systems at /taiga (also /taiga/nsf/delta is bind mounted at /projects) and is accessible on both the Delta and Taiga DTN endpoints. For NCSA & Illinois researchers, Taiga is also mounted on NCSA's HAL and Radiant systems. This storage subsystem has an aggregate performance of 140GB/s and 1PB of its capacity allocated to users of the Delta system. /taiga is a Lustre file system running DDN Exascaler software.
Hardware:
DDN SFA400NVXE (Quantity: 2), each unit contains
- 4 x SS9012 enclosures
- NVME for metadata and small files
DDN SFA18XE (Quantity: 1) *coming soon*, each unit contains
- 10 x SS9012 enclosures
- NVME for for metadata and small files
| Info | ||
|---|---|---|
| ||
A "module reset" in a job script will populate $WORK and $SCRATCH environment variables automatically, or you may set them as WORK=/projects/<account>/$USER , SCRATCH=/scratch/<account>/$USER . |
File System | Quota | Snapshots | Purged | Key Features |
|---|---|---|---|---|
HOME (/u) | 25GB. 400,000 files per user. | No/TBA | No | Area for software, scripts, job files, etc. NOT intended as a source/destination for I/O during jobs |
WORK (/projects) | 500 GB. Up to 1-25 TB by allocation request | No/TBA | No | Area for shared data for a project, common data sets, software, results, etc. |
SCRATCH (/scratch) | 1000 GB. Up to 1-100 TB by allocation request. | No | Yes; files older than 30-days (access time) | Area for computation, largest allocations, where I/O from jobs should occur |
| /tmp | 0.74-1.50 TB shared or dedicated depending on node usage by job(s), no quotas in place | No | After each job | Locally attached disk for fast small file IO. |
quota usage
The quota command allows you to view your use of the file systems and use by your projects. Below is a sample output for a person "user" who is in two projects: aaaa, and bbbb. The home directory quota does not depend on which project group the file is written with.
| Code Block | ||||
|---|---|---|---|---|
| ||||
<user>@dt-login01 ~]$ quota Quota usage for user <user>: ------------------------------------------------------------------------------------------- | Directory Path | User | User | User | User | User | User | | | Block| Soft | Hard | File | Soft | Hard | | | Used | Quota| Limit | Used | Quota | Limit| -------------------------------------------------------------------------------------- | /u/<user> | 20k | 25G | 27.5G | 5 | 300000 | 330000 | -------------------------------------------------------------------------------------- Quota usage for groups user <user> is a member of: ------------------------------------------------------------------------------------- | Directory Path | Group | Group | Group | Group | Group | Group | | | Block | Soft | Hard | File | Soft | Hard | | | Used | Quota | Limit | Used | Quota | Limit | ------------------------------------------------------------------------------------------- | /projects/aaaa | 8k | 500G | 550G | 2 | 300000 | 330000 | | /projects/bbbb | 24k | 500G | 550G | 6 | 300000 | 330000 | | /scratch/aaaa | 8k | 552G | 607.2G| 2 | 500000 | 550000 | | /scratch/bbbb | 24k | 9.766T| 10.74T| 6 | 500000 | 550000 | ------------------------------------------------------------------------------------------ |
File System Dependency Specification for Jobs
We request that jobs specify file system or systems being used in order for us to respond to resource availability issues. We assume that all jobs depend on the HOME file system.
Table of Slurm Feature/constraint labels
| File system | Feature/constraint label | Note |
|---|---|---|
| WORK (/projects) | projects | |
| SCRACH (/scratch) | scratch | |
| IME (/ime) | ime | depends on scratch |
| TAIGA (/taiga) | taiga |
The Slurm constraint specifier and slurm Feature attribute for jobs are used to add file system dependencies to a job.
Slurm Feature Specification
For already submitted and pending (PD) jobs, please use the Slurm Feature attribute as follows:
| Code Block |
|---|
$ scontrol update job=JOBID Features="feature1&feature2" |
For example, to add scratch and ime Features to an already submitted job:
| Code Block |
|---|
$ scontrol update job=713210 Features="scratch&ime" |
To verify the setting:
| Code Block |
|---|
$ scontrol show job 713210 | grep Feature Features=scratch&ime DelayBoot=00:00:00 |
Slurm constraint Specification
To add Slurm job constraint attributes when submitting a job with sbatch (or with srun as a command line argument) use the following:
| Code Block |
|---|
#SBATCH --constraint="constraint1&constraint2.." |
For example, to add scratch and ime constraints to when submitting a job:
| Code Block |
|---|
#SBATCH --constraint="scratch&ime" |
To verify the setting:
| Code Block |
|---|
$ scontrol show job 713267 | grep Feature Features=scratch&ime DelayBoot=00:00:00 |
Accessing the System
Direct Access Anchor login_nodes login_nodes
| login_nodes | |
| login_nodes |
Direct access to the Delta login nodes is via ssh using your NCSA username, password and NCS Duo MFA. Please see NCSA Allocation and Account Management page for links to NCSA Identity and NCSA Duo services. The login nodes provide access to the CPU and GPU resources on Delta.
| login node hostname | example usage with ssh |
|---|---|
| dt-login01.delta.ncsa.illinois.edu | ssh -Y username@dt-login01.delta.ncsa.illinois.edu |
| dt-login02.delta.ncsa.illinois.edu | ssh -l username dt-login02.delta.ncsa.illinois.edu |
login.delta.ncsa.illinois.edu (round robin DNS name for the set of login nodes) | ssh username@login.delta.ncsa.illinois.edu |
Please see NCSA Allocation and Account Management for the steps to change your NCSA password for direct access and set up NCSA DUO. Please contact help@ncsa.illinois.edu for assistance if you do not know your NCSA username.
Use of ssh-key pairs is disabled for general use. Please contact NCSA Help at help@ncsa.illinois.edu for key-pair use by Gateway allocations.
| Info | ||
|---|---|---|
| ||
tmux is available on the login nodes to maintain persistent sessions. See the tmux man page for more information. Use the targeted login hostnames (dt-login01 or dt-login02) to attach to the login node where you started tmux after making note of the hostname. Avoid the round-robin hostname when using tmux. |
Citizenship
You share Delta with thousands of other users , and what you do on the system affects others. Exercise good citizenship to ensure that your activity does not adversely impact the system and the research community with whom you share it. Here are some rules of thumb:
- Don’t run production jobs on the login nodes (very short time debug tests are fine)
- Don’t stress filesystems with known-harmful access patterns (many thousands of small files in a single directory)
- submit an informative help-desk ticket including loaded modules (module list) and stdout/stderr messages
Managing and Transferring Files
File Systems
Each user has a home directory, $HOME, located at /u/$USER.
For example, a user (with username auser) who has an allocated project with a local project serial code abcd will see the following entries in their $HOME and entries in the project and scratch file systems. To determine the mapping of ACCESS project to local project please use the accounts command.
Directory access changes can be made using the facl command. Contact help@ncsa.illinois.edu if you need assistance with enabling access to specific users and projects.
| Code Block | ||
|---|---|---|
| ||
$ ls -ld /u/$USER drwxrwx---+ 12 root root 12345 Feb 21 11:54 /u/$USER $ ls -ld /projects/abcd drwxrws---+ 45 root delta_abcd 4096 Feb 21 11:54 /projects/abcd $ ls -l /projects/abcd total 0 drwxrws---+ 2 auser delta_abcd 6 Feb 21 11:54 auser drwxrws---+ 2 buser delta_abcd 6 Feb 21 11:54 buser ... $ ls -ld /scratch/abcd drwxrws---+ 45 root delta_abcd 4096 Feb 21 11:54 /scratch/abcd $ ls -l /scratch/abcd total 0 drwxrws---+ 2 auser delta_abcd 6 Feb 21 11:54 auser drwxrws---+ 2 buser delta_abcd 6 Feb 21 11:54 buser ... |
To avoid issues when file systems become unstable or non-responsive, we recommend not putting symbolic links from $HOME to the project and scratch spaces.
| Info | ||
|---|---|---|
| ||
The high performance ssd storage (740GB cpu, 1.5TB gpu) is available in /tmp (unique to each node and job–not a shared filesystem) and may contain less than the expected free space if the node(s) are running multiple jobs. Codes that need to perform i/o to many small files should target /tmp on each node of the job and save results to other filesystems before the job ends. |
Transferring your Files
To transfer files to and from the Delta system :
| Info | ||
|---|---|---|
| ||
Many GUI applications that support ssh/scp/sftp will work with DUO. A good first step is to use the interactive (not stored/saved) password option with those apps. The interactive login should present you with the 1st password prompt (your kerberos password) followed by the 2nd password prompt for DUO (push to device or passcode from DUO app). |
- scp - to be used for small to modest transfers to avoid impacting the usability of the Delta login node (login.delta.ncsa.illinois.edu).
- rsync - to be used for small to modest transfers to avoid impacting the usability of the Delta login node.
- https://campuscluster.illinois.edu/resources/docs/storage-and-data-guide/ (scp, sftp, rsync)
Globus - to be used for large data transfers.
Info title Upgrade your Globus Connect Personal Upgrade to at least version 3.2.0 before Dec 12, 2022. See: https://docs.globus.org/ca-update-2022/#notice
- Use the Delta collection "NCSA Delta".
-

- Please see the following documentation on using Globus Online
Sharing Files with Collaborators
Building Software
The Delta programming environment supports the GNU, AMD (AOCC), Intel and NVIDIA HPC compilers. Support for the HPE/Cray Programming environment is forthcoming.
Modules provide access to the compiler + MPI environment.
The default environment includes the GCC 11.2.0 compiler + OpenMPI with support for cuda and gdrcopy. nvcc is in the cuda module and is loaded by default.
AMD recommended compiler flags for GNU, AOCC, and Intel compilers for Milan processors can be found in the AMD Compiler Options Quick Reference Guide for Epyc 7xx3 processors.
View file name Compiler Options Quick Ref Guide for AMD EPYC 7xx3 Series Processors.pdf height 250
Serial
To build (compile and link) a serial program in Fortran, C, and C++:
| gcc | aocc | nvhpc |
|---|---|---|
| gfortran myprog.f gcc myprog.c g++ myprog.cc | flang myprog.f clang myprog.c clang myprog.cc | nvfortran myprog.f nvc myprog.c nvc++ myprog.cc |
MPI
To build (compile and link) a MPI program in Fortran, C, and C++:
| MPI Implementation | modulefiles for MPI/Compiler | Build Commands | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
OpenMPI | aocc/3.2.0 openmpi |
|
OpenMP
To build an OpenMP program, use the -fopenmp / -mp option:
| gcc | aocc | nvhpc |
|---|---|---|
| gfortran -fopenmp myprog.f gcc -fopenmp myprog.c g++ -fopenmp myprog.cc | flang -fopenmp myprog.f clang -fopenmp myprog.c clang -fopenmp myprog.cc | nvfortran -mp myprog.f nvc -mp myprog.c nvc++ -mp myprog.cc |
Hybrid MPI/OpenMP
To build an MPI/OpenMP hybrid program, use the -fopenmp / -mp option with the MPI compiling commands:
| GCC | PGI/NVHPC | |
|---|---|---|
| mpif77 -fopenmp myprog.f mpif90 -fopenmp myprog.f90 mpicc -fopenmp myprog.c mpic++ -fopenmp myprog.cc | mpif77 -mp myprog.f mpif90 -mp myprog.f90 mpicc -mp myprog.c mpic++ -mp myprog.cc |
Cray xthi.c sample code
Document - XC Series User Application Placement Guide CLE6..0UP01 S-2496 | HPE Support
This code can be compiled using the methods show above. The code appears in some of the batch script examples below to demonstrate core placement options.
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
#define _GNU_SOURCE
#include <stdio.h>
#include <unistd.h>
#include <string.h>
#include <sched.h>
#include <mpi.h>
#include <omp.h>
/* Borrowed from util-linux-2.13-pre7/schedutils/taskset.c */
static char *cpuset_to_cstr(cpu_set_t *mask, char *str)
{
char *ptr = str;
int i, j, entry_made = 0;
for (i = 0; i < CPU_SETSIZE; i++) {
if (CPU_ISSET(i, mask)) {
int run = 0;
entry_made = 1;
for (j = i + 1; j < CPU_SETSIZE; j++) {
if (CPU_ISSET(j, mask)) run++;
else break;
}
if (!run)
sprintf(ptr, "%d,", i);
else if (run == 1) {
sprintf(ptr, "%d,%d,", i, i + 1);
i++;
} else {
sprintf(ptr, "%d-%d,", i, i + run);
i += run;
}
while (*ptr != 0) ptr++;
}
}
ptr -= entry_made;
*ptr = 0;
return(str);
}
int main(int argc, char *argv[])
{
int rank, thread;
cpu_set_t coremask;
char clbuf[7 * CPU_SETSIZE], hnbuf[64];
MPI_Init(&argc, &argv);
MPI_Comm_rank(MPI_COMM_WORLD, &rank);
memset(clbuf, 0, sizeof(clbuf));
memset(hnbuf, 0, sizeof(hnbuf));
(void)gethostname(hnbuf, sizeof(hnbuf));
#pragma omp parallel private(thread, coremask, clbuf)
{
thread = omp_get_thread_num();
(void)sched_getaffinity(0, sizeof(coremask), &coremask);
cpuset_to_cstr(&coremask, clbuf);
#pragma omp barrier
printf("Hello from rank %d, thread %d, on %s. (core affinity = %s)\n",
rank, thread, hnbuf, clbuf);
}
MPI_Finalize();
return(0);
} |
A version of xthi is also available from ORNL
| Code Block |
|---|
% git clone https://github.com/olcf/XC30-Training/blob/master/affinity/Xthi.c |
OpenACC
To build an OpenACC program, use the -acc option and the -mp option for multi-threaded:
| NON-MULTITHREADED | MULTITHREADED | |
|---|---|---|
| nvfortran -acc myprog.f nvc -acc myprog.c nvc++ -acc myprog.cc | nvfortran -acc -mp myprog.f nvc -acc -mp myprog.c nvc++ -acc -mp myprog.cc |
CUDA
Cuda compilers (nvcc) are included in the cuda module which is loaded by default under modtree/gpu. For the cuda fortran compiler and other Nvidia development tools, load the "nvhpc" module.
| Code Block | ||
|---|---|---|
| ||
[arnoldg@dt-login03 namd]$ nv nvaccelerror nvidia-bug-report.sh nvlink nvaccelinfo nvidia-cuda-mps-control nv-nsight-cu nvc nvidia-cuda-mps-server nv-nsight-cu-cli nvc++ nvidia-debugdump nvprepro nvcc nvidia-modprobe nvprof nvcpuid nvidia-persistenced nvprune nvcudainit nvidia-powerd nvsize nvdecode nvidia-settings nvunzip nvdisasm nvidia-sleep.sh nvvp nvextract nvidia-smi nvzip nvfortran nvidia-xconfig |
See also: https://developer.nvidia.com/hpc-sdk
HIP / ROCM (AMD MI100)
To access the development environment for the gpuMI100x8 partition, start a job on the node with srun or sbatch. Then set your PATH to prefix /opt/rocm/bin where the HIP and ROCM tools are installed. A sample batch script to obtain an xterm is shown along with setting the path on the compute node:
| Code Block | ||
|---|---|---|
| ||
#!/bin/bash -x MYACCOUNT=$1 GPUS=--gpus-per-node=1 PARTITION=gpuMI100x8-interactive srun --tasks-per-node=1 --nodes=1 --cpus-per-task=1 \ --mem=16g \ --partition=$PARTITION \ --time=00:30:00 \ --account=$MYACCOUNT \ $GPUS --x11 \ xterm |
| Code Block | ||
|---|---|---|
| ||
[arnoldg@gpud01 bin]$ export PATH=/opt/rocm/bin:$PATH [arnoldg@gpud01 bin]$ hipcc No Arguments passed, exiting ... [arnoldg@gpud01 bin]$ |
See also: https://developer.amd.com/resources/rocm-learning-center/fundamentals-of-hip-programming/ , https://rocmdocs.amd.com/en/latest/
Software
Delta software is provisioned, when possible, using spack to produce modules for use via the lmod based module system. Select NVIDIA NGC containers are made available (see the container section below) and are periodically updated from the NVIDIA NGC site. An automated list of available software can be found on the ACCESS website.
modules/lmod
| Anchor | ||||
|---|---|---|---|---|
|
Delta provides two sets of modules and a variety of compilers in each set. The default environment is modtree/gpu which loads a recent version of gnu compilers , the openmpi implementation of MPI, and cuda. The environment with gpu support will build binaries that run on both the gpu nodes (with cuda) and cpu nodes (potentially with warning messages because those nodes lack cuda drivers). For situations where the same version of software is to be deployed on both gpu and cpu nodes but with separate builds, the modtree/cpu environment provides the same default compiler and MPI but without cuda. Use module spider package_name to search for software in lmod and see the steps to load it for your environment.
| module (lmod) command | example | ||
|---|---|---|---|
module list (display the currently loaded modules) |
| ||
module load <package_name> (loads a package or metamodule such as modtree/gpu or netcdf-c) |
| ||
module spider <package_name> (finds modules and displays the ways to load them) module -r spider "regular expression" |
|
see also: User Guide for Lmod
Please open a service request ticket by sending email to help@ncsa.illinois.edu for help with software not currently installed on the Delta system. For single user or single project use cases the preference is for the user to use the spack software package manager to install software locally against the system spack installation as documented <here>. Delta support staff are available to provide limited assistance. For general installation requests the Delta project office will review requests for broad use and installation effort.
Python
On Delta, you may install your own python software stacks as needed. There are a couple choices when customizing your python setup. You may use any of these methods with any of the python versions or instances described below (or you may install your own python versions):
- pip3 : pip3 install --user <python_package>
- useful when you need just 1 python environment per python version or instance
- venv (python virtual environment)
- can name environments (metadata) and have multiple environments per python version or instance
- conda environments
- similar to venv but with more flexibility , see this comparison (1/2 way down the page)
| Info | ||
|---|---|---|
| ||
The Nvidia NGC containers on Delta provide optimized python frameworks built for Delta's A100 and A40 gpus. Delta staff recommend using an NGC container when possible with the gpu nodes (or use the anaconda3_gpu module described later). |
The default gcc (latest version) programming environment for either modtree/cpu or modtree/gpu contains:
Anaconda
anaconda3_cpu
Use python from the anaconda3_cpu module if you need some of the modules provided by Anaconda in your python workflow. See the "managing environments" section of the Conda getting started guide to learn how to customize Conda for your workflow and add extra python modules to your environment. We recommend starting with anaconda3_cpu for modtree/cpu and the cpu nodes, do not use this module with gpus, use anaconda3_gpu instead.
| Warning | ||
|---|---|---|
| ||
If you use anaconda with NGC containers, take care to use the python from the container and not the python from anaconda or one of its environments. The container's python should be 1st in $PATH. You may --bind the anaconda directory or other paths into the container so that you can start your conda environments, but with the container's python (/usr/bin/python). |
| Info | ||
|---|---|---|
| ||
https://repo.anaconda.com/archive/ contains previous Anaconda versions. The bundles are not small, but using one from Anaconda would ensure that you get software that was built to work together at a point in time. If you require an older version of a python lib/module, we suggest looking back in time at the Anaconda site. |
| Code Block | ||
|---|---|---|
| ||
$ module load modtree/cpu $ module load gcc anaconda3_cpu $ which conda /sw/external/python/anaconda3_cpu/conda $ module list Currently Loaded Modules: 1) cue-login-env/1.0 6) libfabric/1.14.0 11) ucx/1.11.2 2) default 7) lustre/2.14.0_ddn23 12) openmpi/4.1.2 3) gcc/11.2.0 8) openssh/8.0p1 13) modtree/cpu 4) knem/1.1.4 9) pmix/3.2.3 14) anaconda3_cpu/4.13.0 5) libevent/2.1.8 10) rdma-core/32.0 |
List of modules in anaconda3_cpu
The current list of modules available in anaconda3_cpu is shown via "conda list", including tensorflow, pytorch, etc:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
# packages in environment at /sw/external/python/anaconda3_cpu: # Name Version Build Channel _ipyw_jlab_nb_ext_conf 0.1.0 py39h06a4308_1 _libgcc_mutex 0.1 main _openmp_mutex 4.5 1_gnu absl-py 1.1.0 pypi_0 pypi aiobotocore 2.3.3 pypi_0 pypi aiohttp 3.8.1 py39h7f8727e_1 aioitertools 0.10.0 pypi_0 pypi aiosignal 1.2.0 pyhd3eb1b0_0 alabaster 0.7.12 pyhd3eb1b0_0 anaconda 2022.05 py39_0 anaconda-client 1.9.0 py39h06a4308_0 anaconda-navigator 2.1.4 py39h06a4308_0 anaconda-project 0.10.2 pyhd3eb1b0_0 anyio 3.5.0 py39h06a4308_0 appdirs 1.4.4 pyhd3eb1b0_0 argon2-cffi 21.3.0 pyhd3eb1b0_0 argon2-cffi-bindings 21.2.0 py39h7f8727e_0 arrow 1.2.2 pyhd3eb1b0_0 astroid 2.6.6 py39h06a4308_0 astropy 5.0.4 py39hce1f21e_0 asttokens 2.0.5 pyhd3eb1b0_0 astunparse 1.6.3 pypi_0 pypi async-timeout 4.0.1 pyhd3eb1b0_0 atomicwrites 1.4.0 py_0 attrs 21.4.0 pyhd3eb1b0_0 automat 20.2.0 py_0 autopep8 1.6.0 pyhd3eb1b0_0 awscli 1.25.14 pypi_0 pypi babel 2.9.1 pyhd3eb1b0_0 backcall 0.2.0 pyhd3eb1b0_0 backports 1.1 pyhd3eb1b0_0 backports.functools_lru_cache 1.6.4 pyhd3eb1b0_0 backports.tempfile 1.0 pyhd3eb1b0_1 backports.weakref 1.0.post1 py_1 bcrypt 3.2.0 py39he8ac12f_0 beautifulsoup4 4.11.1 py39h06a4308_0 binaryornot 0.4.4 pyhd3eb1b0_1 bitarray 2.4.1 py39h7f8727e_0 bkcharts 0.2 py39h06a4308_0 black 19.10b0 py_0 blas 1.0 mkl bleach 4.1.0 pyhd3eb1b0_0 blosc 1.21.0 h8c45485_0 bokeh 2.4.2 py39h06a4308_0 boto3 1.21.32 pyhd3eb1b0_0 botocore 1.24.21 pypi_0 pypi bottleneck 1.3.4 py39hce1f21e_0 brotli 1.0.9 he6710b0_2 brotlipy 0.7.0 py39h27cfd23_1003 brunsli 0.1 h2531618_0 bzip2 1.0.8 h7b6447c_0 c-ares 1.18.1 h7f8727e_0 ca-certificates 2022.3.29 h06a4308_1 cachetools 4.2.2 pyhd3eb1b0_0 certifi 2021.10.8 py39h06a4308_2 cffi 1.15.0 py39hd667e15_1 cfitsio 3.470 hf0d0db6_6 chardet 4.0.0 py39h06a4308_1003 charls 2.2.0 h2531618_0 charset-normalizer 2.0.4 pyhd3eb1b0_0 click 8.0.4 py39h06a4308_0 cloudpickle 2.0.0 pyhd3eb1b0_0 clyent 1.2.2 py39h06a4308_1 colorama 0.4.4 pyhd3eb1b0_0 colorcet 2.0.6 pyhd3eb1b0_0 conda 4.13.0 py39h06a4308_0 conda-build 3.21.8 py39h06a4308_2 conda-content-trust 0.1.1 pyhd3eb1b0_0 conda-env 2.6.0 1 conda-pack 0.6.0 pyhd3eb1b0_0 conda-package-handling 1.8.1 py39h7f8727e_0 conda-repo-cli 1.0.4 pyhd3eb1b0_0 conda-token 0.3.0 pyhd3eb1b0_0 conda-verify 3.4.2 py_1 constantly 15.1.0 pyh2b92418_0 cookiecutter 1.7.3 pyhd3eb1b0_0 cpuonly 2.0 0 pytorch-nightly cryptography 3.4.8 py39hd23ed53_0 cssselect 1.1.0 pyhd3eb1b0_0 curl 7.82.0 h7f8727e_0 cycler 0.11.0 pyhd3eb1b0_0 cython 0.29.28 py39h295c915_0 cytoolz 0.11.0 py39h27cfd23_0 daal4py 2021.5.0 py39h78b71dc_0 dal 2021.5.1 h06a4308_803 dask 2022.2.1 pyhd3eb1b0_0 dask-core 2022.2.1 pyhd3eb1b0_0 dataclasses 0.8 pyh6d0b6a4_7 datashader 0.13.0 pyhd3eb1b0_1 datashape 0.5.4 py39h06a4308_1 dbus 1.13.18 hb2f20db_0 debugpy 1.5.1 py39h295c915_0 decorator 5.1.1 pyhd3eb1b0_0 defusedxml 0.7.1 pyhd3eb1b0_0 diff-match-patch 20200713 pyhd3eb1b0_0 dill 0.3.5.1 pypi_0 pypi distributed 2022.2.1 pyhd3eb1b0_0 docutils 0.16 pypi_0 pypi entrypoints 0.4 py39h06a4308_0 et_xmlfile 1.1.0 py39h06a4308_0 etils 0.7.1 pypi_0 pypi executing 0.8.3 pyhd3eb1b0_0 expat 2.4.4 h295c915_0 ffmpeg 4.2.2 h20bf706_0 filelock 3.6.0 pyhd3eb1b0_0 flake8 3.9.2 pyhd3eb1b0_0 flask 1.1.2 pyhd3eb1b0_0 flatbuffers 1.12 pypi_0 pypi fontconfig 2.13.1 h6c09931_0 fonttools 4.25.0 pyhd3eb1b0_0 freetype 2.11.0 h70c0345_0 frozenlist 1.2.0 py39h7f8727e_0 fsspec 2022.5.0 pypi_0 pypi funcx 1.0.2 pypi_0 pypi funcx-common 0.0.15 pypi_0 pypi future 0.18.2 py39h06a4308_1 gast 0.4.0 pypi_0 pypi gensim 4.1.2 py39h295c915_0 giflib 5.2.1 h7b6447c_0 glib 2.69.1 h4ff587b_1 glob2 0.7 pyhd3eb1b0_0 globus-cli 3.8.0 pypi_0 pypi globus-sdk 3.11.0 pypi_0 pypi gmp 6.2.1 h2531618_2 gmpy2 2.1.2 py39heeb90bb_0 gnutls 3.6.15 he1e5248_0 google-api-core 1.25.1 pyhd3eb1b0_0 google-auth 1.33.0 pyhd3eb1b0_0 google-auth-oauthlib 0.4.6 pypi_0 pypi google-cloud-core 1.7.1 pyhd3eb1b0_0 google-cloud-storage 1.31.0 py_0 google-crc32c 1.1.2 py39h27cfd23_0 google-pasta 0.2.0 pypi_0 pypi google-resumable-media 1.3.1 pyhd3eb1b0_1 googleapis-common-protos 1.53.0 py39h06a4308_0 greenlet 1.1.1 py39h295c915_0 grpcio 1.42.0 py39hce63b2e_0 gst-plugins-base 1.14.0 h8213a91_2 gstreamer 1.14.0 h28cd5cc_2 gviz-api 1.10.0 pypi_0 pypi h5py 3.6.0 py39ha0f2276_0 hdf5 1.10.6 hb1b8bf9_0 heapdict 1.0.1 pyhd3eb1b0_0 holoviews 1.14.8 pyhd3eb1b0_0 hvplot 0.7.3 pyhd3eb1b0_1 hyperlink 21.0.0 pyhd3eb1b0_0 icu 58.2 he6710b0_3 idna 3.3 pyhd3eb1b0_0 imagecodecs 2021.8.26 py39h4cda21f_0 imageio 2.9.0 pyhd3eb1b0_0 imagesize 1.3.0 pyhd3eb1b0_0 importlib-metadata 4.11.3 py39h06a4308_0 importlib-resources 5.9.0 pypi_0 pypi importlib_metadata 4.11.3 hd3eb1b0_0 incremental 21.3.0 pyhd3eb1b0_0 inflection 0.5.1 py39h06a4308_0 iniconfig 1.1.1 pyhd3eb1b0_0 intake 0.6.5 pyhd3eb1b0_0 intel-openmp 2021.4.0 h06a4308_3561 intervaltree 3.1.0 pyhd3eb1b0_0 ipykernel 6.9.1 py39h06a4308_0 ipython 8.2.0 py39h06a4308_0 ipython_genutils 0.2.0 pyhd3eb1b0_1 ipywidgets 7.6.5 pyhd3eb1b0_1 isort 5.9.3 pyhd3eb1b0_0 itemadapter 0.3.0 pyhd3eb1b0_0 itemloaders 1.0.4 pyhd3eb1b0_1 itsdangerous 2.0.1 pyhd3eb1b0_0 jax 0.3.16 pypi_0 pypi jaxlib 0.3.15 pypi_0 pypi jdcal 1.4.1 pyhd3eb1b0_0 jedi 0.18.1 py39h06a4308_1 jeepney 0.7.1 pyhd3eb1b0_0 jinja2 2.11.3 pyhd3eb1b0_0 jinja2-time 0.2.0 pyhd3eb1b0_3 jmespath 0.10.0 pyhd3eb1b0_0 joblib 1.1.0 pyhd3eb1b0_0 jpeg 9e h7f8727e_0 jq 1.6 h27cfd23_1000 json5 0.9.6 pyhd3eb1b0_0 jsonschema 4.4.0 py39h06a4308_0 jupyter 1.0.0 py39h06a4308_7 jupyter_client 6.1.12 pyhd3eb1b0_0 jupyter_console 6.4.0 pyhd3eb1b0_0 jupyter_core 4.9.2 py39h06a4308_0 jupyter_server 1.13.5 pyhd3eb1b0_0 jupyterlab 3.3.2 pyhd3eb1b0_0 jupyterlab_pygments 0.1.2 py_0 jupyterlab_server 2.10.3 pyhd3eb1b0_1 jupyterlab_widgets 1.0.0 pyhd3eb1b0_1 jxrlib 1.1 h7b6447c_2 keras 2.9.0 pypi_0 pypi keras-preprocessing 1.1.2 pypi_0 pypi keyring 23.4.0 py39h06a4308_0 kiwisolver 1.3.2 py39h295c915_0 krb5 1.19.2 hac12032_0 lame 3.100 h7b6447c_0 lazy-object-proxy 1.6.0 py39h27cfd23_0 lcms2 2.12 h3be6417_0 ld_impl_linux-64 2.35.1 h7274673_9 lerc 3.0 h295c915_0 libaec 1.0.4 he6710b0_1 libarchive 3.4.2 h62408e4_0 libclang 14.0.1 pypi_0 pypi libcrc32c 1.1.1 he6710b0_2 libcurl 7.82.0 h0b77cf5_0 libdeflate 1.8 h7f8727e_5 libedit 3.1.20210910 h7f8727e_0 libev 4.33 h7f8727e_1 libffi 3.3 he6710b0_2 libgcc-ng 9.3.0 h5101ec6_17 libgfortran-ng 7.5.0 ha8ba4b0_17 libgfortran4 7.5.0 ha8ba4b0_17 libgomp 9.3.0 h5101ec6_17 libidn2 2.3.2 h7f8727e_0 liblief 0.11.5 h295c915_1 libllvm11 11.1.0 h3826bc1_1 libnghttp2 1.46.0 hce63b2e_0 libopus 1.3.1 h7b6447c_0 libpng 1.6.37 hbc83047_0 libprotobuf 3.19.1 h4ff587b_0 libsodium 1.0.18 h7b6447c_0 libspatialindex 1.9.3 h2531618_0 libssh2 1.10.0 h8f2d780_0 libstdcxx-ng 9.3.0 hd4cf53a_17 libtasn1 4.16.0 h27cfd23_0 libtiff 4.2.0 h85742a9_0 libunistring 0.9.10 h27cfd23_0 libuuid 1.0.3 h7f8727e_2 libvpx 1.7.0 h439df22_0 libwebp 1.2.2 h55f646e_0 libwebp-base 1.2.2 h7f8727e_0 libxcb 1.14 h7b6447c_0 libxml2 2.9.12 h03d6c58_0 libxslt 1.1.34 hc22bd24_0 libzopfli 1.0.3 he6710b0_0 llvmlite 0.38.0 py39h4ff587b_0 locket 0.2.1 py39h06a4308_2 lxml 4.8.0 py39h1f438cf_0 lz4-c 1.9.3 h295c915_1 lzo 2.10 h7b6447c_2 markdown 3.3.4 py39h06a4308_0 markupsafe 2.0.1 py39h27cfd23_0 matplotlib 3.5.1 py39h06a4308_1 matplotlib-base 3.5.1 py39ha18d171_1 matplotlib-inline 0.1.2 pyhd3eb1b0_2 mccabe 0.6.1 py39h06a4308_1 mistune 0.8.4 py39h27cfd23_1000 mkl 2021.4.0 h06a4308_640 mkl-service 2.4.0 py39h7f8727e_0 mkl_fft 1.3.1 py39hd3c417c_0 mkl_random 1.2.2 py39h51133e4_0 mock 4.0.3 pyhd3eb1b0_0 mpc 1.1.0 h10f8cd9_1 mpfr 4.0.2 hb69a4c5_1 mpi 1.0 mpich mpich 3.3.2 hc856adb_0 mpmath 1.2.1 py39h06a4308_0 msgpack-python 1.0.2 py39hff7bd54_1 multidict 5.2.0 py39h7f8727e_2 multipledispatch 0.6.0 py39h06a4308_0 munkres 1.1.4 py_0 mypy_extensions 0.4.3 py39h06a4308_1 navigator-updater 0.2.1 py39_1 nbclassic 0.3.5 pyhd3eb1b0_0 nbclient 0.5.13 py39h06a4308_0 nbconvert 6.4.4 py39h06a4308_0 nbformat 5.3.0 py39h06a4308_0 ncurses 6.3 h7f8727e_2 nest-asyncio 1.5.5 py39h06a4308_0 nettle 3.7.3 hbbd107a_1 networkx 2.7.1 pyhd3eb1b0_0 nltk 3.7 pyhd3eb1b0_0 nose 1.3.7 pyhd3eb1b0_1008 notebook 6.4.8 py39h06a4308_0 numba 0.55.1 py39h51133e4_0 numexpr 2.8.1 py39h6abb31d_0 numpy 1.21.5 py39he7a7128_1 numpy-base 1.21.5 py39hf524024_1 numpydoc 1.2 pyhd3eb1b0_0 oauthlib 3.2.0 pypi_0 pypi olefile 0.46 pyhd3eb1b0_0 oniguruma 6.9.7.1 h27cfd23_0 openh264 2.1.1 h4ff587b_0 openjpeg 2.4.0 h3ad879b_0 openpyxl 3.0.9 pyhd3eb1b0_0 openssl 1.1.1n h7f8727e_0 opt-einsum 3.3.0 pypi_0 pypi packaging 21.3 pyhd3eb1b0_0 pandas 1.4.2 py39h295c915_0 pandocfilters 1.5.0 pyhd3eb1b0_0 panel 0.13.0 py39h06a4308_0 param 1.12.0 pyhd3eb1b0_0 parsel 1.6.0 py39h06a4308_0 parso 0.8.3 pyhd3eb1b0_0 partd 1.2.0 pyhd3eb1b0_1 patchelf 0.13 h295c915_0 pathspec 0.7.0 py_0 patsy 0.5.2 py39h06a4308_1 pcre 8.45 h295c915_0 pep8 1.7.1 py39h06a4308_0 pexpect 4.8.0 pyhd3eb1b0_3 pickleshare 0.7.5 pyhd3eb1b0_1003 pillow 9.0.1 py39h22f2fdc_0 pip 21.2.4 py39h06a4308_0 pkginfo 1.8.2 pyhd3eb1b0_0 plotly 5.6.0 pyhd3eb1b0_0 pluggy 1.0.0 py39h06a4308_1 poyo 0.5.0 pyhd3eb1b0_0 prometheus_client 0.13.1 pyhd3eb1b0_0 prompt-toolkit 3.0.20 pyhd3eb1b0_0 prompt_toolkit 3.0.20 hd3eb1b0_0 protego 0.1.16 py_0 protobuf 3.19.1 py39h295c915_0 psutil 5.8.0 py39h27cfd23_1 ptyprocess 0.7.0 pyhd3eb1b0_2 pure_eval 0.2.2 pyhd3eb1b0_0 py 1.11.0 pyhd3eb1b0_0 py-lief 0.11.5 py39h295c915_1 pyasn1 0.4.8 pyhd3eb1b0_0 pyasn1-modules 0.2.8 py_0 pycodestyle 2.7.0 pyhd3eb1b0_0 pycosat 0.6.3 py39h27cfd23_0 pycparser 2.21 pyhd3eb1b0_0 pyct 0.4.6 py39h06a4308_0 pycurl 7.44.1 py39h8f2d780_1 pydantic 1.10.2 pypi_0 pypi pydispatcher 2.0.5 py39h06a4308_2 pydocstyle 6.1.1 pyhd3eb1b0_0 pyerfa 2.0.0 py39h27cfd23_0 pyflakes 2.3.1 pyhd3eb1b0_0 pygments 2.11.2 pyhd3eb1b0_0 pyhamcrest 2.0.2 pyhd3eb1b0_2 pyjwt 2.1.0 py39h06a4308_0 pylint 2.9.6 py39h06a4308_1 pyls-spyder 0.4.0 pyhd3eb1b0_0 pyodbc 4.0.32 py39h295c915_1 pyopenssl 21.0.0 pyhd3eb1b0_1 pyparsing 3.0.4 pyhd3eb1b0_0 pyqt 5.9.2 py39h2531618_6 pyrsistent 0.18.0 py39heee7806_0 pysocks 1.7.1 py39h06a4308_0 pytables 3.6.1 py39h77479fe_1 pytest 7.1.1 py39h06a4308_0 python 3.9.12 h12debd9_0 python-dateutil 2.8.2 pyhd3eb1b0_0 python-fastjsonschema 2.15.1 pyhd3eb1b0_0 python-libarchive-c 2.9 pyhd3eb1b0_1 python-lsp-black 1.0.0 pyhd3eb1b0_0 python-lsp-jsonrpc 1.0.0 pyhd3eb1b0_0 python-lsp-server 1.2.4 pyhd3eb1b0_0 python-slugify 5.0.2 pyhd3eb1b0_0 python-snappy 0.6.0 py39h2531618_3 pytorch 1.13.0.dev20220620 py3.9_cpu_0 pytorch-nightly pytorch-mutex 1.0 cpu pytorch-nightly pytz 2021.3 pyhd3eb1b0_0 pyviz_comms 2.0.2 pyhd3eb1b0_0 pywavelets 1.3.0 py39h7f8727e_0 pyxdg 0.27 pyhd3eb1b0_0 pyyaml 5.4.1 pypi_0 pypi pyzmq 22.3.0 py39h295c915_2 qdarkstyle 3.0.2 pyhd3eb1b0_0 qstylizer 0.1.10 pyhd3eb1b0_0 qt 5.9.7 h5867ecd_1 qtawesome 1.0.3 pyhd3eb1b0_0 qtconsole 5.3.0 pyhd3eb1b0_0 qtpy 2.0.1 pyhd3eb1b0_0 queuelib 1.5.0 py39h06a4308_0 readline 8.1.2 h7f8727e_1 regex 2022.3.15 py39h7f8727e_0 requests 2.27.1 pyhd3eb1b0_0 requests-file 1.5.1 pyhd3eb1b0_0 requests-oauthlib 1.3.1 pypi_0 pypi ripgrep 12.1.1 0 rope 0.22.0 pyhd3eb1b0_0 rsa 4.7.2 pyhd3eb1b0_1 rtree 0.9.7 py39h06a4308_1 ruamel_yaml 0.15.100 py39h27cfd23_0 s3fs 2022.5.0 pypi_0 pypi s3transfer 0.6.0 pypi_0 pypi scikit-image 0.19.2 py39h51133e4_0 scikit-learn 1.0.2 py39h51133e4_1 scikit-learn-intelex 2021.5.0 py39h06a4308_0 scipy 1.7.3 py39hc147768_0 scrapy 2.6.1 py39h06a4308_0 seaborn 0.11.2 pyhd3eb1b0_0 secretstorage 3.3.1 py39h06a4308_0 send2trash 1.8.0 pyhd3eb1b0_1 service_identity 18.1.0 pyhd3eb1b0_1 setuptools 61.2.0 py39h06a4308_0 sip 4.19.13 py39h295c915_0 six 1.16.0 pyhd3eb1b0_1 smart_open 5.1.0 pyhd3eb1b0_0 snappy 1.1.9 h295c915_0 sniffio 1.2.0 py39h06a4308_1 snowballstemmer 2.2.0 pyhd3eb1b0_0 sortedcollections 2.1.0 pyhd3eb1b0_0 sortedcontainers 2.4.0 pyhd3eb1b0_0 soupsieve 2.3.1 pyhd3eb1b0_0 sphinx 4.4.0 pyhd3eb1b0_0 sphinxcontrib-applehelp 1.0.2 pyhd3eb1b0_0 sphinxcontrib-devhelp 1.0.2 pyhd3eb1b0_0 sphinxcontrib-htmlhelp 2.0.0 pyhd3eb1b0_0 sphinxcontrib-jsmath 1.0.1 pyhd3eb1b0_0 sphinxcontrib-qthelp 1.0.3 pyhd3eb1b0_0 sphinxcontrib-serializinghtml 1.1.5 pyhd3eb1b0_0 spyder 5.1.5 py39h06a4308_1 spyder-kernels 2.1.3 py39h06a4308_0 sqlalchemy 1.4.32 py39h7f8727e_0 sqlite 3.38.2 hc218d9a_0 stack_data 0.2.0 pyhd3eb1b0_0 statsmodels 0.13.2 py39h7f8727e_0 sympy 1.10.1 py39h06a4308_0 tabulate 0.8.9 py39h06a4308_0 tbb 2021.5.0 hd09550d_0 tbb4py 2021.5.0 py39hd09550d_0 tblib 1.7.0 pyhd3eb1b0_0 tenacity 8.0.1 py39h06a4308_0 tensorboard 2.9.1 pypi_0 pypi tensorboard-data-server 0.6.1 pypi_0 pypi tensorboard-plugin-profile 2.8.0 pypi_0 pypi tensorboard-plugin-wit 1.8.1 pypi_0 pypi tensorflow 2.9.1 pypi_0 pypi tensorflow-estimator 2.9.0 pypi_0 pypi tensorflow-io-gcs-filesystem 0.26.0 pypi_0 pypi termcolor 1.1.0 pypi_0 pypi terminado 0.13.1 py39h06a4308_0 testpath 0.5.0 pyhd3eb1b0_0 text-unidecode 1.3 pyhd3eb1b0_0 textdistance 4.2.1 pyhd3eb1b0_0 threadpoolctl 2.2.0 pyh0d69192_0 three-merge 0.1.1 pyhd3eb1b0_0 tifffile 2021.7.2 pyhd3eb1b0_2 tinycss 0.4 pyhd3eb1b0_1002 tk 8.6.11 h1ccaba5_0 tldextract 3.2.0 pyhd3eb1b0_0 toml 0.10.2 pyhd3eb1b0_0 tomli 1.2.2 pyhd3eb1b0_0 toolz 0.11.2 pyhd3eb1b0_0 torchaudio 0.13.0.dev20220621 py39_cpu pytorch-nightly torchvision 0.14.0.dev20220621 py39_cpu pytorch-nightly tornado 6.1 py39h27cfd23_0 tqdm 4.64.0 py39h06a4308_0 traitlets 5.1.1 pyhd3eb1b0_0 twisted 22.2.0 py39h7f8727e_0 typed-ast 1.4.3 py39h7f8727e_1 typing-extensions 4.1.1 hd3eb1b0_0 typing_extensions 4.1.1 pyh06a4308_0 tzdata 2022a hda174b7_0 ujson 5.1.0 py39h295c915_0 unidecode 1.2.0 pyhd3eb1b0_0 unixodbc 2.3.9 h7b6447c_0 urllib3 1.26.9 py39h06a4308_0 w3lib 1.21.0 pyhd3eb1b0_0 watchdog 2.1.6 py39h06a4308_0 wcwidth 0.2.5 pyhd3eb1b0_0 webencodings 0.5.1 py39h06a4308_1 websocket-client 0.58.0 py39h06a4308_4 websockets 10.3 pypi_0 pypi werkzeug 2.0.3 pyhd3eb1b0_0 wget 1.21.3 h0b77cf5_0 wheel 0.37.1 pyhd3eb1b0_0 widgetsnbextension 3.5.2 py39h06a4308_0 wrapt 1.12.1 py39he8ac12f_1 wurlitzer 3.0.2 py39h06a4308_0 x264 1!157.20191217 h7b6447c_0 xarray 0.20.1 pyhd3eb1b0_1 xlrd 2.0.1 pyhd3eb1b0_0 xlsxwriter 3.0.3 pyhd3eb1b0_0 xz 5.2.5 h7b6447c_0 yaml 0.2.5 h7b6447c_0 yapf 0.31.0 pyhd3eb1b0_0 yarl 1.6.3 py39h27cfd23_0 zeromq 4.3.4 h2531618_0 zfp 0.5.5 h295c915_6 zict 2.0.0 pyhd3eb1b0_0 zipp 3.7.0 pyhd3eb1b0_0 zlib 1.2.12 h7f8727e_2 zope 1.0 py39h06a4308_1 zope.interface 5.4.0 py39h7f8727e_0 zstd 1.4.9 haebb681_0 |
anaconda3_gpu
Similar to the setup for anaconda, we have a gpu version of anaconda3 (module load anaconda3_gpu) and have installed pytorch and tensorflow cuda aware python modules into this version. You may use this module when working with the gpu nodes. See conda list after loading the module to review what is already installed. As with anaconda3_cpu, let Delta staff know if there are generally useful modules you would like us to try to install for the broader community. A sample tensorflow test script:
| Code Block | ||||
|---|---|---|---|---|
| ||||
#!/bin/bash
#SBATCH --mem=64g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=2 # <- match to OMP_NUM_THREADS
#SBATCH --partition=gpuA100x4-interactive
#SBATCH --time=00:10:00
#SBATCH --account=YOUR_ACCOUNT-delta-gpu
#SBATCH --job-name=tf_anaconda
### GPU options ###
#SBATCH --gpus-per-node=1
#SBATCH --gpus-per-task=1
#SBATCH --gpu-bind=verbose,per_task:1
###SBATCH --gpu-bind=none # <- or closest
module purge # drop modules and explicitly load the ones needed
# (good job metadata and reproducibility)
module load anaconda3_gpu
module list # job documentation and metadata
echo "job is starting on `hostname`"
which python3
conda list tensorflow
srun python3 \
tf_gpu.py
exit |
Jupyter notebooks
The Detla Open OnDemand portal provides an easier way to start a Jupyter notebook. Please see OpenOnDemand to access the portal.
The jupyter notebook executables are in your $PATH after loading the anaconda3 module. Don't run jupyter on the shared login nodes. Instead, follow these steps to attach a jupyter notebook running on a compute node to your local web browser:
1) Start a jupyter job via srun and note the hostname (you pick the port number for --port). |
Use the 2nd URL in step 3. Note the internal hostname in the cluster for step 2. When using a container with a gpu node, run the container's jupyter-notebook:
In step 3 to start the notebook in your browser, replace http://hostname:8888/ with http://127.0.0.1:8991/ ( the port number you selected with --port= ) You may not see the job hostname when running with a container, find it with squeue:
Then specifu the host your job is using in the next step (gpua045 for example ). | |||||||||||||||
| 2) From your local desktop or laptop create an ssh tunnel to the compute node via a login node of delta. |
Authenticate with your login and 2-factor as usual. | |||||||||||||||
3) Paste the 2nd URL (containing 127.0.0.1:port_number and the token string) from step 1 into your browser and you will be connected to the jupyter instance running on your compute node of Delta.
|
|
Python (a recent or latest version)
If you do not need all of the extra modules provided by Anaconda, use the basic python installation under the gcc module. You can add modules via "pip3 install --user <modulename>", setup virtual environments, and customize as needed for your workflow but starting from a smaller installed base of python than Anaconda.
| Code Block | ||
|---|---|---|
| ||
$ module load gcc python $ which python /sw/spack/delta-2022-03/apps/python/3.10.4-gcc-11.2.0-3cjjp6w/bin/python $ module list Currently Loaded Modules: 1) modtree/gpu 3) gcc/11.2.0 5) ucx/1.11.2 7) python/3.10.4 2) default 4) cuda/11.6.1 6) openmpi/4.1.2 |
This is the list of modules available in the python from "pip3 list":
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
Package Version ------------------ --------- certifi 2021.10.8 cffi 1.15.0 charset-normalizer 2.0.12 click 8.1.2 cryptography 36.0.2 globus-cli 3.4.0 globus-sdk 3.5.0 idna 3.3 jmespath 0.10.0 pip 22.0.4 pycparser 2.21 PyJWT 2.3.0 requests 2.27.1 setuptools 58.1.0 urllib3 1.26.9 |
Launching Applications
- Launching One Serial Application
- Launching One Multi-Threaded Application
- Launching One MPI Application
- Launching One Hybrid (MPI+Threads) Application
- More Than One Serial Application in the Same Job
- MPI Applications One at a Time
- More than One MPI Application Running Concurrently
- More than One OpenMP Application Running Concurrently
Running Jobs
Job Accounting
The charge unit for Delta is the Service Unit (SU). This corresponds to the equivalent use of one compute core utilizing less than or equal to 2G of memory for one hour, or 1 GPU or fractional GPU using less than the corresponding amount of memory or cores for 1 hour (see table below). Keep in mind that your charges are based on the resources that are reserved for your job and don't necessarily reflect how the resources are used. Charges are based on either the number of cores or the fraction of the memory requested, whichever is larger. The minimum charge for any job is 1 SU.
Node Type | Service Unit Equivalence | |||
|---|---|---|---|---|
| Cores | GPU Fraction | Host Memory | ||
| CPU Node | 11 | N/A | 2 GB | |
GPU Node | Quad A100 | 16 | 1 A100 | 64 62.5 GB |
| Quad A40 | 16 | 1 A40 | 64 62.5 GB | |
| 8-way A100 | 16 | 1 A100 | 250 GB | |
| 8-way MI100 | 16 | 1 MI100 | 250 GB | |
Please note that a weighting factor will discount the charge for the reduced-precision A40 nodes, as well as the novel AMD MI100 based node - this will be documented through the ACCESS SU converter.
Local Account Charging
Use the accounts command to list the accounts available for charging. CPU and GPU resources will have individual charge names. For example in the following, abcd-delta-cpu and abcd-delta-gpu are available for user gbauer to use for the CPU and GPU resources.
| Code Block | ||
|---|---|---|
| ||
$ accounts availableProject Slurm accountsSummary for userUser gbauer'kingda': abcd-delta-cpu my_prefix my project abcd-delta-gpu my_prefix my project Project Description Balance (Hours) Deposited (Hours) -------------- --------------------------- ----------------- ------------------- bbka-delta-gpu ncsa/delta staff allocation 5000000 5000000 bbka-delta-cpu ncsa/delta staff allocation 100000000 100000000 |
Job Accounting Considerations
- A node-exclusive job that runs on a compute node for one hour will be charged 128 SUs (128 cores x 1 hour)
- A node-exclusive job that runs on a 4-way GPU node for one hour will be charge 4 SUs (4 GPU x 1 hour)
- A node-exclusive job that runs on a 8-way GPU node for one hour will be charge 8 SUs (8 GPU x 1 hour)A shared job that runs on an A100 node will be charged for the fractional usage of the A100.
Accessing the Compute Nodes
Delta implements the Slurm batch environment to manage access to the compute nodes. Use the Slurm commands to run batch jobs or for interactive access to compute nodes. See: https://slurm.schedmd.com/quickstart.html for an introduction to Slurm. There are two ways to access compute nodes on Delta.
Batch jobs can be used to access compute nodes. Slurm provides a convenient direct way to submit batch jobs. See https://slurm.schedmd.com/heterogeneous_jobs.html#submitting for details.
Sample Slurm batch job scripts are provided in the Job Scripts section below.
Direct ssh access to a compute node in a running batch job from a dt-loginNN node is enabled, once the job has started.
| Code Block |
|---|
$ squeue --job jobid
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12345 cpu bash gbauer R 0:17 1 cn001 |
Then in a terminal session:
| Code Block |
|---|
$ ssh cn001 cn001.delta.internal.ncsa.edu (172.28.22.64) OS: RedHat 8.4 HW: HPE CPU: 128x RAM: 252 GB Site: mgmt Role: compute $ |
Scheduler
For information, consult:
https://slurm.schedmd.com/quickstart.html
slurm quick reference guide View file name slurm_summary.pdf height 250
Partitions (Queues)
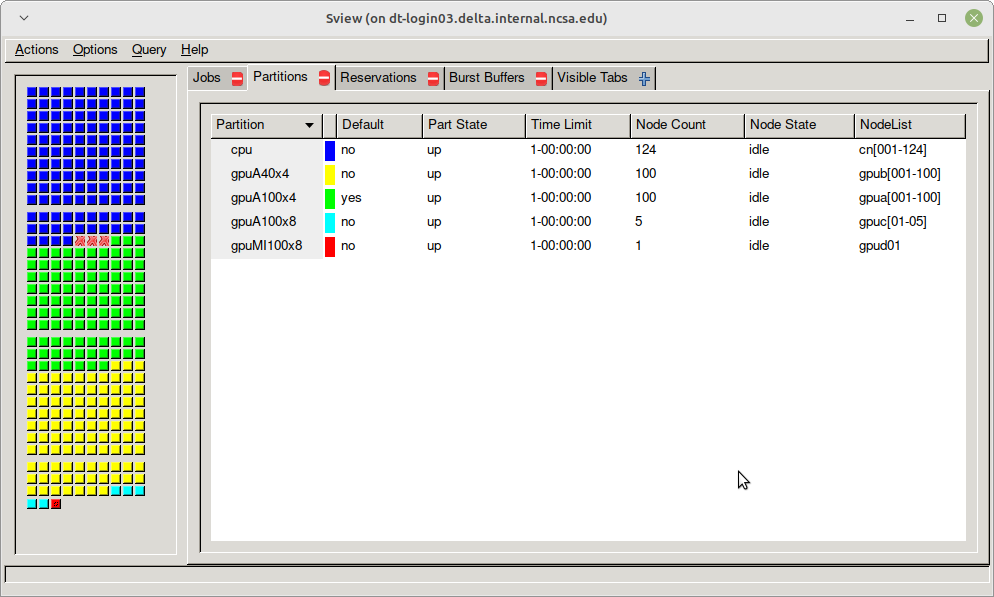
Table. Delta Production Early Access Period Partitions/Queues
Partition/Queue | Node Type | Max Nodes per Job | Max Duration | Max Running in Queue/user* | Charge Factor |
|---|---|---|---|---|---|
cpu | CPU | TBD | 24 hr / 48 hr | 8,448 cores | 1.0 |
| cpu-interactive | CPU | TBD | 30 min | in total | 2.0 |
gpuA100x4 gpuA100x4* (asterisk indicates this is the default queue, but submit jobs to gpuA100x4) | quad A100 | TBD | 24 hr / 48 hr | 3,200 cores and 200 gpus | 1.0 |
| gpuA100x4-interactive | quad-A100 | TBD | 30 min | in total | 2.0 |
| gpuA100x8 | octa-A100 | TBD | 24 hr / 48 hr | TBD | 2.0 |
| gpuA100x8-interactive | octa-A100 | TBD | 30 min | TBD | 4.0 |
| gpuA40x4 | quad-A40 | TBD | 24 hr / 48 hr | 3,200 cores and 200 gpus | 0.6 |
| gpuA40x4-interactive | quad-A40 | TBD | 30 min | in total | 1.2 |
| gpuMI100x8 | octa-MI100 | TBD | 24 hr / 48 hr | TBD | 1.5 |
| gpuMI100x8-interactive | octa-MI100 | TBD | 30 min | TBD | 3.0 |
sview view of slurm partitions
Node Policies
Node-sharing is the default for jobs. Node-exclusive mode can be obtained by specifying all the consumable resources for that node type or adding the following Slurm options:
--exclusive --mem=0
GPU NVIDIA MIG (GPU slicing) for the A100 will be supported at a future date.
Pre-emptive jobs will be supported at a future date.
Interactive Sessions
Interactive sessions can be implemented in several ways depending on what is needed.
To start up a bash shell terminal on a cpu or gpu node
- single core with 1GB of memory, with one task on a cpu node
| Code Block |
|---|
srun --account=account_name --partition=cpu-interactive \ --nodes=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=16g \ --pty bash |
- single core with 20GB of memory, with one task on a A40 gpu node
| Code Block |
|---|
srun --account=account_name --partition=gpuA40x4-interactive \ --nodes=1 --gpus-per-node=1 --tasks=1 \ --tasks-per-node=1 --cpus-per-task=1 --mem=20g \ --pty bash |
| Info | ||
|---|---|---|
| ||
Since interactive jobs are already a child process of srun, one cannot srun applications from within them. Use mpirun to launch mpi jobs from within an interactive job. Within standard batch jobs submitted via sbatch, use srun to launch MPI codes. |
Interactive X11 Support
To run an X11 based application on a compute node in an interactive session, the use of the --x11 switch with srun is needed. For example, to run a single core job that uses 1g of memory with X11 (in this case an xterm) do the following:
| Code Block |
|---|
srun -A abcd-delta-cpu --partition=cpu-interactive \ --nodes=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=16g \ --x11 xterm |
File System Dependency Specification for Jobs
Please see the FileSystemDependencySpecificationforJobs section on setting job file system dependencies for jobs.
Jobs that do not specify a dependency on the WORK(/projects) and SCRATCH (/scratch) will be assumed to depend only on the HOME (/u) file system.
Sample Job Scripts Anchor Job Scripts Job Scripts
| Job Scripts | |
| Job Scripts |
Serial jobs
Code Block language bash title serial example script $ cat job.slurm #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=1 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=myjobtest #SBATCH --time=00:10:00 # hh:mm:ss for the job #SBATCH --constraint="scratch" ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module reset # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) # $WORK and $SCRATCH are now set module load python # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" srun python3 myprog.pyMPI
Code Block language bash title mpi example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=2 #SBATCH --ntasks-per-node=32 #SBATCH --cpus-per-task=1 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=mympi #SBATCH --time=00:10:00 # hh:mm:ss for the job #SBATCH --constraint="scratch" ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module reset # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) # $WORK and $SCRATCH are now set module load gcc/11.2.0 openmpi # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" srun osu_reduceOpenMP
Code Block language bash title openmp example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=32 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=myopenmp #SBATCH --time=00:10:00 # hh:mm:ss for the job #SBATCH --constraint="scratch" ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module reset # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) # $WORK and $SCRATCH are now set module load gcc/11.2.0 # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" export OMP_NUM_THREADS=32 srun stream_gccHybrid (MPI + OpenMP or MPI+X)
Code Block language bash title mpi+x example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 #SBATCH --cpus-per-task=4 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=mympi+x #SBATCH --time=00:10:00 # hh:mm:ss for the job #SBATCH --constraint="scratch" ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module reset # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) # $WORK and $SCRATCH are now set module load gcc/11.2.0 openmpi # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" export OMP_NUM_THREADS=4 srun xthi4 gpus together on a compute node
Code Block language bash title 4 gpus on a compute node collapse true #!/bin/bash #SBATCH --job-name="a.out_symmetric" #SBATCH --output="a.out.%j.%N.out" #SBATCH --partition=gpuA100x4 #SBATCH --mem=220G #SBATCH --nodes=1 #SBATCH --ntasks-per-node=4 #SBATCH --cpus-per-task=16 # spread out to use 1 core per numa #SBATCH --constraint="scratch" #SBATCH --gpus-per-node=4 #SBATCH --gpu-bind=closest # select a cpu close to gpu on pci bus topology #SBATCH --account=bbjw-delta-gpu #SBATCH --exclusive # dedicated node for this job #SBATCH --no-requeue #SBATCH -t 04:00:00 export OMP_NUM_THREADS=1 # if code is not multithreaded, otherwise set to 8 or 16 srun -N 1 -n 4 ./a.out > myjob.out
- Parametric / Array / HTC jobs
Job Management
Batch jobs are submitted through a job script (as in the examples above) using the sbatch command. Job scripts generally start with a series of SLURM directives that describe requirements of the job such as number of nodes, wall time required, etc… to the batch system/scheduler (SLURM directives can also be specified as options on the sbatch command line; command line options take precedence over those in the script). The rest of the batch script consists of user commands.
The syntax for sbatch is:
sbatch [list of sbatch options] script_name
Refer to the sbatch man page for detailed information on the options.
squeue/scontrol/sinfo
Commands that display batch job and partition information .
| SLURM EXAMPLE COMMAND | DESCRIPTION |
|---|---|
| squeue -a | List the status of all jobs on the system. |
| squeue -u $USER | List the status of all your jobs in the batch system. |
| squeue -j JobID | List nodes allocated to a running job in addition to basic information.. |
| scontrol show job JobID | List detailed information on a particular job. |
| sinfo -a | List summary information on all the partition. |
See the manual (man) pages for other available options.
Job Status
NODELIST(REASON)
MaxGRESPerAccount - a user has exceeded the number of cores or gpus allotted per user or project for a given partition.
Useful Batch Job Environment Variables
| Anchor | ||||
|---|---|---|---|---|
|
DESCRIPTION | SLURM ENVIRONMENT VARIABLE | DETAIL DESCRIPTION |
|---|---|---|
| JobID | $SLURM_JOB_ID | Job identifier assigned to the job |
| Job Submission Directory | $SLURM_SUBMIT_DIR | By default, jobs start in the directory that the job was submitted from. So the "cd $SLURM_SUBMIT_DIR" command is not needed. |
| Machine(node) list | $SLURM_NODELIST | variable name that contains the list of nodes assigned to the batch job |
| Array JobID | $SLURM_ARRAY_JOB_ID $SLURM_ARRAY_TASK_ID | each member of a job array is assigned a unique identifier |
See the sbatch man page for additional environment variables available.
srun
The srun command initiates an interactive job on the compute nodes.
For example, the following command:
srun -A account_name --time=00:30:00 --nodes=1 --ntasks-per-node=64 \
--mem=16g --pty /bin/bash
will run an interactive job in the default queue with a wall clock limit of 30 minutes, using one node and 16 cores per node. You can also use other sbatch options such as those documented above.
After you enter the command, you will have to wait for SLURM to start the job. As with any job, your interactive job will wait in the queue until the specified number of nodes is available. If you specify a small number of nodes for smaller amounts of time, the wait should be shorter because your job will backfill among larger jobs. You will see something like this:
srun: job 123456 queued and waiting for resources
Once the job starts, you will see:
srun: job 123456 has been allocated resources
and will be presented with an interactive shell prompt on the launch node. At this point, you can use the appropriate command to start your program.
When you are done with your work, you can use the exit command to end the job.
scancel
The scancel command deletes a queued job or terminates a running job.
- scancel JobID deletes/terminates a job.
Refunds
Refunds are considered, when appropriate, for jobs that failed due to circumstances beyond user control.
Projects wishing to request a refund should email help@ncsa.illinois.edu. Please include the batch job ids and the standard error and output files produced by the job(s).
Visualization
Delta A40 nodes support NVIDIA raytracing hardware.
- describe visualization capabilities & software.
- how to establish VNC/DVC/remote desktop
Containers
Apptainer (formerly Singularity)
Container support on Delta is provided by Apptainer/Singularity.
Docker images can be converted to Singularity sif format via the singularity pull command. Commands can be run from within a container using singularity run command (or apptainer run).
If you encounter quota issues with Apptainer/Singularity caching in ~/.singularity , the environment variable SINGULARITY_CACHEDIR can be used to use a different location such as a scratch space.
Your $HOME is automatically available from containers run via Apptainer/Singularity. You can "pip3 install --user" against a container's python, setup virtualenv's or similar while useing a containerized application. Just run the container's /bin/bash to get a Singularity> prompt. Here's an srun example of that with tensorflow:
| Code Block | ||||
|---|---|---|---|---|
| ||||
$ srun \ --mem=32g \ --nodes=1 \ --ntasks-per-node=1 \ --cpus-per-task=1 \ --partition=gpuA100x4-interactive \ --account=bbka-delta-gpu \ --gpus-per-node=1 \ --gpus-per-task=1 \ --gpu-bind=verbose,per_task:1 \ --pty \ apptainer run --nv \ /sw/external/NGC/tensorflow:22.06-tf2-py3 /bin/bash # job starts ... Singularity> hostname gpua068.delta.internal.ncsa.edu Singularity> which python # the python in the container /usr/bin/python Singularity> python --version Python 3.8.10 Singularity> |
NVIDIA NGC Containers
Delta provides NVIDIA NGC Docker containers that we have pre-built with Singularity. Look for the latest binary containers in /sw/external/NGC/ . The containers are used as shown in the sample scripts below:
| Code Block | ||||
|---|---|---|---|---|
| ||||
#!/bin/bash
#SBATCH --mem=64g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=64 # <- match to OMP_NUM_THREADS, 64 requests whole node
#SBATCH --partition=gpuA100x4 # <- one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8
#SBATCH --account=bbka-delta-gpu
#SBATCH --job-name=pytorchNGC
### GPU options ###
#SBATCH --gpus-per-node=1
#SBATCH --gpus-per-task=1
#SBATCH --gpu-bind=verbose,per_task:1
module reset # drop modules and explicitly load the ones needed
# (good job metadata and reproducibility)
# $WORK and $SCRATCH are now set
module list # job documentation and metadata
echo "job is starting on `hostname`"
# run the container binary with arguments: python3 <program.py>
apptainer run --nv \
/sw/external/NGC/pytorch:22.02-py3 python3 tensor_gpu.py |
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
#!/bin/bash
#SBATCH --mem=64g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=64 # <- match to OMP_NUM_THREADS
#SBATCH --partition=gpuA100x4 # <- one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8
#SBATCH --account=bbka-delta-gpu
#SBATCH --job-name=tfNGC
### GPU options ###
#SBATCH --gpus-per-node=1
#SBATCH --gpus-per-task=1
#SBATCH --gpu-bind=verbose,per_task:1
module reset # drop modules and explicitly load the ones needed
# (good job metadata and reproducibility)
# $WORK and $SCRATCH are now set
module list # job documentation and metadata
echo "job is starting on `hostname`"
# run the container binary with arguments: python3 <program.py>
singularity run --nv \
/sw/external/NGC/tensorflow:22.06-tf2-py3 python3 \
tf_matmul.py |
Container list (as of March, 2022)
| Code Block | ||
|---|---|---|
| ||
caffe:20.03-py3 caffe2:18.08-py3 catalog.txt cntk:18.08-py3 cp2k_v9.1.0.sif cuquantum-appliance_22.03-cirq.sif digits:21.09-tensorflow-py3 gromacs_2022.1.sif hpc-benchmarks:21.4-hpl lammps:patch_4May2022 matlab:r2021b mxnet:21.09-py3 mxnet_22.08-py3.sif namd_2.13-multinode.sif namd_3.0-alpha11.sif paraview_egl-py3-5.9.0.sif pytorch:22.02-py3 pytorch_22.07-py3.sif pytorch_22.08-py3.sif tensorflow_19.09-py3.sif tensorflow:22.02-tf1-py3 tensorflow:22.02-tf2-py3 tensorflow_22.05-tf1-py3.sif tensorflow_22.05-tf2-py3.sif tensorflow:22.06-tf1-py3 tensorflow:22.06-tf2-py3 tensorflow_22.07-tf2-py3.sif tensorflow_22.08-tf1-py3.sif tensorflow_22.08-tf2-py3.sif tensorrt:22.02-py3 tensorrt_22.08-py3.sif theano:18.08 torch:18.08-py2 |
see also: https://catalog.ngc.nvidia.com/orgs/nvidia/containers
AMD Infinity Hub containers for MI100
The AMD node in partition gpuMI100x8 (-interactive) will run containers from the AMD Infinity Hub. The Delta team has pre loaded the following containers in /sw/external/MI100 and will retrieve others upon request.
| Code Block | ||
|---|---|---|
| ||
cp2k_8.2.sif gromacs_2021.1.sif lammps_2021.5.14_121.sif milc_c30ed15e1-20210420.sif namd_2.15a2-20211101.sif namd3_3.0a9.sif openmm_7.7.0_49.sif pytorch_rocm5.0_ubuntu18.04_py3.7_pytorch_1.10.0.sif tensorflow_rocm5.0-tf2.7-dev.sif |
A sample batch script for pytorch resembles:
| Code Block | ||||
|---|---|---|---|---|
| ||||
#!/bin/bash
#SBATCH --mem=64g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=1
#SBATCH --partition=gpuMI100x8
#SBATCH --account=bbka-delta-gpu
#SBATCH --job-name=tfAMD
#SBATCH --reservation=amd
#SBATCH --time=00:15:00
### GPU options ###
#SBATCH --gpus-per-node=1
##SBATCH --gpus-per-task=1
##SBATCH --gpu-bind=none # <- or closest
module purge # drop modules and explicitly load the ones needed
# (good job metadata and reproducibility)
module list # job documentation and metadata
echo "job is starting on `hostname`"
# https://apptainer.org/docs/user/1.0/gpu.html#amd-gpus-rocm
# https://pytorch.org/docs/stable/notes/hip.html
time \
apptainer run --rocm \
~arnoldg/delta/AMD/pytorch_rocm5.0_ubuntu18.04_py3.7_pytorch_1.10.0.sif \
python3 tensor_gpu.py
exit |
Other Containers
Extreme-scale Scientific Software Stack (E4S)
The E4S container with GPU (cuda and rocm) support is provided for users of specific ECP packages made available by the E4S project (https://e4s-project.github.io/). The singularity image is available as :
/sw/external/E4S/e4s-gpu-x86_64.sif
To use E4S with NVIDIA GPUs
| Code Block |
|---|
$ srun --account=account_name --partition=gpuA100-interactive \ --nodes=1 --gpus-per-node=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=20g \ --pty bash $ singularity exec --cleanenv /sw/external/E4S/e4s-gpu-x86_64.sif \ /bin/bash --rcfile /etc/bash.bashrc |
The spack package inside of the image will interact with a local spack installation. If ~/.spack directory exists, it might need to be renamed.
More information can be found at https://e4s-project.github.io/download.html
Delta Science Gateway and Open OnDemand
Open OnDemand
The Delta Open OnDemand portal is now available for use. Current supported Interactive apps: Jupyter notebooks.
To connect to the Open OnDemand portal, director a browser to https://openondemand.delta.ncsa.illinois.edu/ and use your NCSA username, password with NCSA Duo with the CILogin page.
Delta Science Gateway
Protected Data (N/A)
Help
For assistance with the use of Delta
ACCESS users can create a ticket via the Help Desk.
- All other users (Illinois allocations, Diversity Allocations, etc) please send email to help@ncsa.illinois.edu.
Acknowledge
To acknowledge the NCSA Delta system in particular, please include the following
This research is part of the Delta research computing project, which is supported by the National Science Foundation (award OCI 2005572), and the State of Illinois. Delta is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications.
How to Acknowledge ACCESS
Acknowledgement for ACCESS Users
References
Supporting documentation resources:
https://www.rcac.purdue.edu/knowledge/anvil
https://nero-docs.stanford.edu/jupyter-slurm.html