Versions Compared
Key
- This line was added.
- This line was removed.
- Formatting was changed.
| Table of Contents |
|---|
Anchor top top
Delta User Guide
| top | |
| top |
Last update: April 7, 2022
Status Updates and Notices
Delta is tentatively scheduled to enter production in Q2 2022.
| Info | ||
|---|---|---|
| ||
| Items in light grey font are in progress and coming soon. Example software or feature not yet implemented. |
Introduction
Delta is a dedicated, eXtreme Science and Engineering Science Discovery Environment (XSEDE) allocated resource designed by HPE and NCSA, delivering a highly capable GPU-focused compute environment for GPU and CPU workloads. Besides offering a mix of standard and reduced precision GPU resources, Delta also offers GPU-dense nodes with both NVIDIA and AMD GPUs. Delta provides high performance node-local SSD scratch filesystems, as well as both standard lustre and relaxed-POSIX parallel filesystems spanning the entire resource.
Delta's CPU nodes are each powered by two 64-core AMD EPYC 7763 ("Milan") processors, with 256 GB of DDR4 memory. The Delta GPU resource has four node types: one with 4 NVIDIA A100 GPUs (40 GB HBM2 RAM each) connected via NVLINK and 1 64-core AMD EPYC 7763 ("Milan") processor, the second with 4 NVIDIA A40 GPUs (48 GB GDDR6 RAM) connected via PCIe 4.0 and 1 64-core AMD EPYC 7763 ("Milan") processor, the third with 8 NVIDIA A100 GPUs in a dual socket AMD EPYC 7763 (128-cores per node) node with 2 TB of DDR4 RAM and NVLINK, and the fourth with 8 AMD MI100 GPUs (32GB HBM2 RAM each) in a dual socket AMD EPYC 7763 (128-cores per node) node with 2 TB of DDR4 RAM and PCIe 4.0.
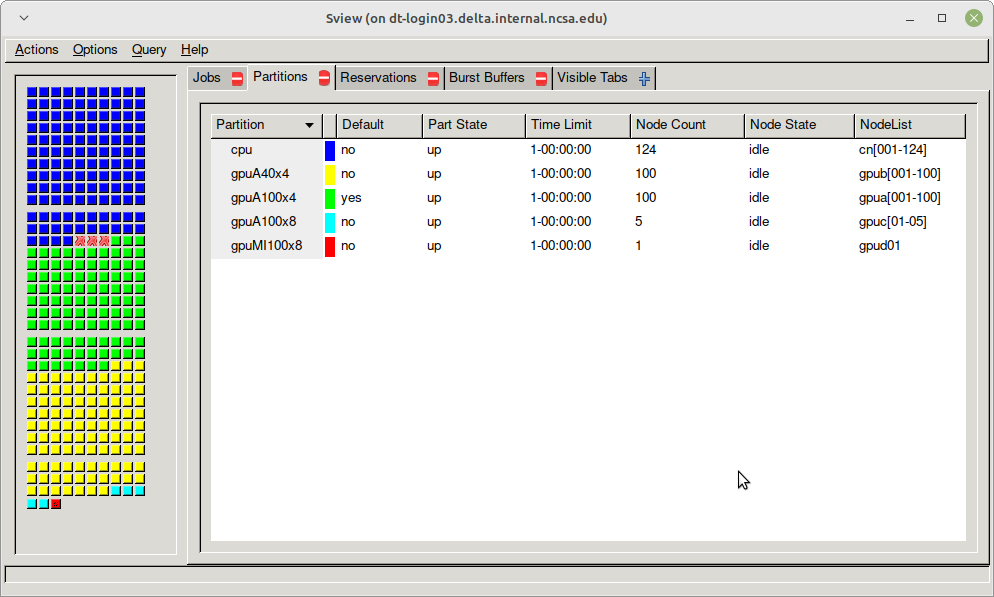
Delta has 124 standard CPU nodes, 100 4-way A100-based GPU nodes, 100 4-way A40-based GPU nodes, 5 8-way A100-based GPU nodes, and 1 8-way MI100-based GPU node. Every Delta node has high-performance node-local SSD storage (740 GB for CPU nodes, 1.5 TB for GPU nodes), and is connected to the 7 PB Lustre parallel filesystem via the high-speed interconnect. The Delta resource uses the SLURM workload manager for job scheduling.
Delta supports the XSEDE core software stack, including remote login, remote computation, data movement, science workflow support, and science gateway support toolkits.
Figure 1. Delta System
Delta is supported by the National Science Foundation under Grant No. OAC-2005572.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
| Delta is now accepting proposals. |
|---|
Account Administration
- For XSEDE projects please use the XSEDE user portal for project and account management.
- Non-XSEDE Account and Project administration is handled by NCSA Identity and NCSA group management tools. For more information please see the NCSA Allocation and Account Management documentation page.
Configuring Your Account
- bash is the default shell, submit a support request to change your default shell
- environment variables: XSEDE CUE, SLURM batch
- using Modules
System Architecture
Delta is designed to help applications transition from CPU-only to GPU or hybrid CPU-GPU codes. Delta has some important architectural features to facilitate new discovery and insight:
- a single processor architecture (AMD) across all node types: CPU and GPU
- support for NVIDIA A100 MIG GPU partitioning allowing for fractional use of the A100s if your workload isn't able to exploit an entire A100 efficiently
- ray tracing hardware support from the NVIDIA A40 GPUs
- 9 large memory (2 TB) nodes
- a low latency and high bandwidth HPE/Cray Slingshot interconnect between compute nodes
- lustre for home, projects and scratch file systems
- support for relaxed and non-posix IO
- shared-node jobs and the single core and single MIG GPU slice
- Resources for persistent services in support of Gateways, Open OnDemand, Data Transport nodes...,
- Unique AMD MI-100 resource
Model Compute Nodes
The Delta compute ecosystem is composed of 5 node types: dual-socket CPU-only compute nodes, single socket 4-way NVIDIA A100 GPU compute nodes, single socket 4-way NVIDIA A40 GPU compute nodes, dual-socket 8-way NVIDIA A100 GPU compute nodes, and a single socket 8-way AMD MI100 GPU compute nodes. The CPU-only and 4-way GPU nodes have 256 GB of RAM per node while the 8-way GPU nodes have 2 TB of RAM. The CPU-only node has 0.74 TB of local storage while all GPU nodes have 1.5 TB of local storage.
Table. CPU Compute Node Specifications
| Specification | Value |
|---|---|
Number of nodes | 124 |
| CPU | AMD EPYC 7763 "Milan" (PCIe Gen4) |
| Sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 0.74 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 4-way NVIDIA A40 GPU Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 100 |
| GPU | NVIDIA A40 |
| GPUs per node | 4 |
| GPU Memory (GB) | 48 DDR6 with ECC |
| CPU | AMD Milan |
| CPU sockets per node | 1 |
Cores per socket | 64 |
| Cores per node | 64 |
Hardware threads per core | 1 |
Hardware threads per node | 64 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
The A40 GPUs are connected via PCIe Gen4 and have the following affinitization to NUMA nodes on the CPU. Note that the relationship between GPU index and NUMA domain are inverse.
Table. 4-way NVIDIA A40 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | SYS | SYS | SYS | SYS | 48-63 | 3 |
| GPU1 | SYS | X | SYS | SYS | SYS | 32-47 | 2 |
| GPU2 | SYS | SYS | X | SYS | SYS | 16-31 | 1 |
| GPU3 | SYS | SYS | SYS | X | PHB | 0-15 | 0 |
| HSN | SYS | SYS | SYS | PHB | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 4-way NVIDIA A100 GPU Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 100 |
| GPU | NVIDIA A100 |
| GPUs per node | 4 |
| GPU Memory (GB) | 40 |
| CPU | AMD Milan |
| CPU sockets per node | 1 |
Cores per socket | 64 |
| Cores per node | 64 |
Hardware threads per core | 1 |
Hardware threads per node | 64 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 256 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 4-way NVIDIA A100 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | NV4 | NV4 | NV4 | SYS | 48-63 | 3 |
| GPU1 | NV4 | X | NV4 | NV4 | SYS | 32-47 | 2 |
| GPU2 | NV4 | NV4 | X | NV4 | SYS | 16-31 | 1 |
| GPU3 | NV4 | NV4 | NV4 | X | PHB | 0-15 | 0 |
| HSN | SYS | SYS | SYS | PHB | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 8-way NVIDIA A100 GPU Large Memory Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 5 |
| GPU | NVIDIA A100 |
| GPUs per node | 8 |
| GPU Memory (GB) | 40 |
| CPU | AMD Milan |
| CPU sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 2,048 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
The AMD CPUs are set for 4 NUMA domains per socket (NPS=4).
Table. 8-way NVIDIA A100 Mapping and GPU-CPU Affinitization
| GPU0 | GPU1 | GPU2 | GPU3 | GPU4 | GPU5 | GPU6 | GPU7 | HSN | CPU Affinity | NUMA Affinity | |
| GPU0 | X | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 48-63 | 3 |
| GPU1 | NV12 | X | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 48-63 | 3 |
| GPU2 | NV12 | NV12 | X | NV12 | NV12 | NV12 | NV12 | NV12 | SYS | 16-31 | 1 |
| GPU3 | NV12 | NV12 | NV12 | X | NV12 | NV12 | NV12 | NV12 | SYS | 16-31 | 1 |
| GPU0 | NV12 | NV12 | NV12 | NV12 | X | NV12 | NV12 | NV12 | SYS | 112-127 | 7 |
| GPU1 | NV12 | NV12 | NV12 | NV12 | NV12 | X | NV12 | NV12 | SYS | 112-127 | 7 |
| GPU2 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | X | NV12 | SYS | 80-95 | 5 |
| GPU3 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | NV12 | X | SYS | 80-95 | 5 |
| HSN | SYS | SYS | SYS | SYS | SYS | SYS | SYS | SYS | X |
Table Legend
X = Self
SYS = Connection traversing PCIe as well as the SMP interconnect between NUMA nodes (e.g., QPI/UPI)
NODE = Connection traversing PCIe as well as the interconnect between PCIe Host Bridges within a NUMA node
PHB = Connection traversing PCIe as well as a PCIe Host Bridge (typically the CPU)
NV# = Connection traversing a bonded set of # NVLinks
Table. 8-way AMD MI100 GPU Large Memory Compute Node Specifications
| Specification | Value |
|---|---|
| Number of nodes | 1 |
| GPU | AMD MI100 |
| GPUs per node | 8 |
| GPU Memory (GB) | 32 |
| CPU | AMD Milan |
| CPU sockets per node | 2 |
Cores per socket | 64 |
| Cores per node | 128 |
Hardware threads per core | 1 |
Hardware threads per node | 128 |
Clock rate (GHz) | ~ 2.45 |
RAM (GB) | 2,048 |
Cache (KB) L1/L2/L3 | 64/512/32768 |
Local storage (TB) | 1.5 TB |
Login Nodes
Login nodes provide interactive support for code compilation.
Specialized Nodes
Delta will support data transfer nodes (serving the "NCSA Delta" Globus Online collection) and nodes in support of other services.
Network
Delta is connected to the NPCF core router & exit infrastructure via two 100Gbps connections, NCSA's 400Gbps+ of WAN connectivity carry traffic to/from users on an optimal peering.
Delta resources are inter-connected with HPE/Cray's 100Gbps/200Gbps SlingShot interconnect.
File Systems
Note: Users of Delta have access to 3 file systems at the time of system launch, a fourth relaxed-POSIX file system will be made available at a later date.
Delta
The Delta storage infrastructure provides users with their $HOME and $SCRATCH areas. These file systems are mounted across all Delta nodes and are accessible on the Delta DTN Endpoints. The aggregate performance of this subsystem is 70GB/s and it has 6PB of usable space. These file systems run Lustre via DDN's ExaScaler 6 stack (Lustre 2.14 based).
Hardware:
DDN SFA7990XE (Quantity: 3), each unit contains
- One additional SS9012 enclosure
- 168 x 16TB SAS Drives
- 7 x 1.92TB SAS SSDs
The $HOME file system has 4 OSTs and is set with a default stripe size of 1.
The $SCRATCH file system has 8 OSTs and has Lustre Progressive File Layout (PFL) enabled which automatically restripes a file as the file grows. The thresholds for PFL restriping for $SCRATCH are
| File size | stripe count |
|---|---|
| 0-32M | 1 OST |
| 32M-512M | 4 OST |
| 512M+ | 8 OST |
Future Hardware:
An additional pool of NVME flash from DDN will be installed in early Spring 2022. This flash will initially be deployed as a tier for "hot" data in scratch. This subsystem will have an aggregate performance of 600GB/s and will have 3PB of capacity. As noted above this subsystem will transition to a relax POSIX namespace file system, communications on that timeline will be announced as updates are available.
Taiga
Taiga is NCSA’s global file system which provides users with their $WORK area. This file system is mounted across all Delta systems at /taiga (also /taiga/nsf/delta is bind mounted at /projects) and is accessible on both the Delta and Taiga DTN endpoints. For NCSA & Illinois researchers, Taiga is also mounted on HAL and Radiant. This storage subsystem has an aggregate performance of 140GB/s and 1PB of its capacity allocated to users of the Delta system. /taiga is a Lustre file system running DDN Exascaler.
Hardware:
DDN SFA400NVXE (Quantity: 2), each unit contains
- 4 x SS9012 enclosures
- NVME for metadata and small files
DDN SFA18XE (Quantity: 1), each unit contains
- 10 x SS9012 enclosures
File System | Quota | Snapshots | Purged | Key Features |
|---|---|---|---|---|
$HOME | 25GB. 400,000 files per user. | No/TBA | No | Area for software, scripts, job files, etc. NOT intended as a source/destination for I/O during jobs |
$WORK | 500 GB. Up to 1-25 TB by allocation request | No/TBA | No | Area for shared data for a project, common data sets, software, results, etc. |
$SCRATCH | 1000 GB. Up to 1-100 TB by allocation request. | No | Yes; files older than 30-days (access time) | Area for computation, largest allocations, where I/O from jobs should occur |
| $LOCAL_SCR namespace mapped to /tmp | shared or dedicated depending on node usage by job(s), no quotas in place | No | After each job | Locally attached disk for fast small file IO. |
Accessing the System
Direct Access Anchor login_nodes login_nodes
| login_nodes | |
| login_nodes |
Direct access to the Delta login nodes is via ssh. The login nodes provide access to the CPU and GPU resources on Delta.
| login node hostname | example usage with ssh |
|---|---|
| dt-login01.delta.ncsa.illinois.edu | ssh -Y username@dt-login01.delta.ncsa.illinois.edu |
| dt-login02.delta.ncsa.illinois.edu | ssh -l username dt-login02.delta.ncsa.illinois.edu |
login.delta.ncsa.illinois.edu (round robin DNS name for the set of login nodes) | ssh username@login.delta.ncsa.illinois.edu |
If needed, XSEDE users can lookup their local username at https://portal.xsede.org/group/xup/accounts. If you need to set a NCSA password for direct access please contact help@ncsa.illinois.edu for assistance.
Use of ssh-key pairs is disabled for general use. Please contact NCSA Help at help@ncsa.illinois.edu for key-pair use by Gateway allocations.
| Info | ||
|---|---|---|
| ||
tmux is available on the login nodes to maintain persistent sessions. See the tmux man page for more information. Use the targeted login hostnames (dt-login01 or dt-login02) to attach to the login node where you started tmux after making note of the hostname. Avoid the round-robin hostname when using tmux. |
XSEDE Single Sign-On Hub
XSEDE users can also access Delta via the XSEDE Single Sign-On Hub.
When reporting a problem to the help desk, please execute the gsissh command with the “-vvv” option and include the verbose output in your problem description.
Once on the XSEDE SSO hub:
| Code Block |
|---|
$ gsissh delta |
or
| Code Block |
|---|
$ gsissh -p 222 delta.ncsa.xsede.org |
The XSEDE SSO with gsi-ssh uses your XSEDE username for login. If that processes is not working, please try using your NCSA username which can be looked up at https://portal.xsede.org/group/xup/accounts (requires XSEDE login).
Citizenship
You share Delta with thousands of other users, and what you do on the system affects others. Exercise good citizenship to ensure that your activity does not adversely impact the system and the research community with whom you share it. Here are some rules of thumb:
- Don’t run production jobs on the login nodes (very short time debug tests are fine)
- Don’t stress filesystems with known-harmful access patterns (many thousands of small files in a single directory)
- submit an informative help-desk ticket including loaded modules (module list) and stdout/stderr messages
Managing and Transferring Files
File Systems
Each user has a home directory, $HOME, located at /u/$USER.
For example, a user (with username auser) who has an allocated project with a local project serial code abcd will see the following entries in their $HOME and entries in the project and scratch file systems. To determine the mapping of XSEDE project to local project please use the accounts command or the userinfo command.
Directory access changes can be made using the facl command. Contact help@ncsa.illinois.edu if you need assistance with enabling access to specific users and projects.
| Code Block | ||
|---|---|---|
| ||
$ ls -ld /u/$USER drwxrwx---+ 12 root root 12345 Feb 21 11:54 /u/$USER $ ls -ld /projects/abcd drwxrws---+ 45 root delta_abcd 4096 Feb 21 11:54 /projects/abcd $ ls -l /projects/abcd total 0 drwxrws---+ 2 auser delta_abcd 6 Feb 21 11:54 auser drwxrws---+ 2 buser delta_abcd 6 Feb 21 11:54 buser ... $ ls -ld /scratch/abcd drwxrws---+ 45 root delta_abcd 4096 Feb 21 11:54 /scratch/abcd $ ls -l /scratch/abcd total 0 drwxrws---+ 2 auser delta_abcd 6 Feb 21 11:54 auser drwxrws---+ 2 buser delta_abcd 6 Feb 21 11:54 buser ... |
To avoid issues when file systems become unstable or non-responsive, we recommend not putting symbolic links from $HOME to the project and scratch spaces.
| Info | ||
|---|---|---|
| ||
The high performance ssd storage (740GB cpu, 1.5TB gpu) is available in /tmp (unique to each node and job–not a shared filesystem) and may contain less than the expected free space if the node(s) are running multiple jobs. Codes that need to perform i/o to many small files should target /tmp on each node of the job and save results to other filesystems before the job ends. |
Transferring your Files
To transfer files to and from the Delta system p
- scp - to be used for small to modest transfers to avoid impacting the usability of the Delta login node.
- rsync - to be used for small to modest transfers to avoid impacting the usability of the Delta login node.
- https://campuscluster.illinois.edu/resources/docs/storage-and-data-guide/ (scp, sftp, rsync)
- Globus - to be used for large data transfers.
- Use the Delta collection "NCSA Delta".
- Please see the following documentation on using Globus Online
Sharing Files with Collaborators
Building Software
The Delta programming environment supports the GNU, AMD (AOCC), Intel and NVIDIA HPC compilers. Support for the HPE/Cray Programming environment is forthcoming.
Modules provide access to the compiler + MPI environment.
The default environment includes the GCC 11.2.0 compiler + OpenMPI with support for cuda and gdrcopy. nvcc is in the cuda module and is loaded by default.
AMD recommended compiler flags for GNU, AOCC, and Intel compilers for Milan processors can be found in the AMD Compiler Options Quick Reference Guide for Epyc 7xx3 processors.
View file name Compiler Options Quick Ref Guide for AMD EPYC 7xx3 Series Processors.pdf height 250
Serial
To build (compile and link) a serial program in Fortran, C, and C++:
| gcc | aocc | nvhpc |
|---|---|---|
| gfortran myprog.f gcc myprog.c g++ myprog.cc | flang myprog.f clang myprog.c clang myprog.cc | nvfortran myprog.f nvc myprog.c nvc++ myprog.cc |
MPI
To build (compile and link) a MPI program in Fortran, C, and C++:
| MPI Implementation | modulefiles for MPI/Compiler | Build Commands | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
OpenMPI | aocc/3.2.0 openmpi |
|
OpenMP
To build an OpenMP program, use the -fopenmp / -mp option:
| gcc | aocc | nvhpc |
|---|---|---|
| gfortran -fopenmp myprog.f gcc -fopenmp myprog.c g++ -fopenmp myprog.cc | flang -fopenmp myprog.f clang -fopenmp myprog.c clang -fopenmp myprog.cc | nvfortran -mp myprog.f nvc -mp myprog.c nvc++ -mp myprog.cc |
Hybrid MPI/OpenMP
To build an MPI/OpenMP hybrid program, use the -fopenmp / -mp option with the MPI compiling commands:
| GCC | PGI/NVHPC | |
|---|---|---|
| mpif77 -fopenmp myprog.f mpif90 -fopenmp myprog.f90 mpicc -fopenmp myprog.c mpic++ -fopenmp myprog.cc | mpif77 -mp myprog.f mpif90 -mp myprog.f90 mpicc -mp myprog.c mpic++ -mp myprog.cc |
Cray xthi.c sample code
Document - XC Series User Application Placement Guide CLE6..0UP01 S-2496 | HPE Support
This code can be compiled using the methods show above. The code appears in some of the batch script examples below to demonstrate core placement options.
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
#define _GNU_SOURCE
#include <stdio.h>
#include <unistd.h>
#include <string.h>
#include <sched.h>
#include <mpi.h>
#include <omp.h>
/* Borrowed from util-linux-2.13-pre7/schedutils/taskset.c */
static char *cpuset_to_cstr(cpu_set_t *mask, char *str)
{
char *ptr = str;
int i, j, entry_made = 0;
for (i = 0; i < CPU_SETSIZE; i++) {
if (CPU_ISSET(i, mask)) {
int run = 0;
entry_made = 1;
for (j = i + 1; j < CPU_SETSIZE; j++) {
if (CPU_ISSET(j, mask)) run++;
else break;
}
if (!run)
sprintf(ptr, "%d,", i);
else if (run == 1) {
sprintf(ptr, "%d,%d,", i, i + 1);
i++;
} else {
sprintf(ptr, "%d-%d,", i, i + run);
i += run;
}
while (*ptr != 0) ptr++;
}
}
ptr -= entry_made;
*ptr = 0;
return(str);
}
int main(int argc, char *argv[])
{
int rank, thread;
cpu_set_t coremask;
char clbuf[7 * CPU_SETSIZE], hnbuf[64];
MPI_Init(&argc, &argv);
MPI_Comm_rank(MPI_COMM_WORLD, &rank);
memset(clbuf, 0, sizeof(clbuf));
memset(hnbuf, 0, sizeof(hnbuf));
(void)gethostname(hnbuf, sizeof(hnbuf));
#pragma omp parallel private(thread, coremask, clbuf)
{
thread = omp_get_thread_num();
(void)sched_getaffinity(0, sizeof(coremask), &coremask);
cpuset_to_cstr(&coremask, clbuf);
#pragma omp barrier
printf("Hello from rank %d, thread %d, on %s. (core affinity = %s)\n",
rank, thread, hnbuf, clbuf);
}
MPI_Finalize();
return(0);
} |
A version of xthi is also available from ORNL
| Code Block |
|---|
% git clone https://github.com/olcf/XC30-Training/blob/master/affinity/Xthi.c |
OpenACC
To build an OpenACC program, use the -acc option and the -mp option for multi-threaded:
| NON-MULTITHREADED | MULTITHREADED | |
|---|---|---|
| nvfortran -acc myprog.f nvc -acc myprog.c nvc++ -acc myprog.cc | nvfortran -acc -mp myprog.f nvc -acc -mp myprog.c nvc++ -acc -mp myprog.cc |
Software
Delta software is provisioned, when possible, using spack to produce modules for use via the lmod based module system. Select NVIDIA NGC containers are made available (see the container section below) and are periodically updated from the NVIDIA NGC site. An automated list of available software can be found on the XSEDE website.
modules/lmod
| Anchor | ||||
|---|---|---|---|---|
|
Delta provides two sets of modules and a variety of compilers in each set. The default environment is modtree/gpu which loads a recent version of gnu compilers , the openmpi implementation of MPI, and cuda. The environment with gpu support will build binaries that run on both the gpu nodes (with cuda) and cpu nodes (potentially with warning messages because those nodes lack cuda drivers). For situations where the same version of software is to be deployed on both gpu and cpu nodes but with separate builds, the modtree/cpu environment provides the same default compiler and MPI but without cuda. Use module spider package_name to search for software in lmod and see the steps to load it for your environment.
| module (lmod) command | example | ||
|---|---|---|---|
module list (display the currently loaded modules) |
| ||
module load <package_name> (loads a package or metamodule such as modtree/gpu or netcdf-c) |
| ||
module spider <package_name> (finds modules and displays the ways to load them) module -r spider "regular expression" |
|
see also: User Guide for Lmod
Please open a service request ticket by sending email to help@ncsa.illinois.edu for help with software not currently installed on the Delta system. For single user or single project use cases the preference is for the user to use the spack software package manager to install software locally against the system spack installation as documented <here>. Delta support staff are available to provide limited assistance. For general installation requests the Delta project office will review requests for broad use and installation effort.
Python
On Delta, you may install your own python software stacks as needed. The default gcc (latest version) programming environment for either modtree/cpu or modtree/gpu contains:
Anaconda3
Use python from the anaconda3 module if you need some of the modules provided by Anaconda in your python workflow. See the "managing environments" section of the Conda getting started guide to learn how to customize Conda for your workflow and add extra python modules to your environment.
| Code Block | ||
|---|---|---|
| ||
$ module load gcc anaconda3 $ which conda /sw/spack/delta-2022-03/apps/anaconda3/2021.05-gcc-11.2.0-ievmolz/condabin/conda $ module list Currently Loaded Modules: 1) modtree/gpu 3) gcc/11.2.0 5) ucx/1.11.2 7) anaconda3/2021.05 2) default 4) cuda/11.6.1 6) openmpi/4.1.2 |
List of modules in anaconda3
The current list of modules available in anaconda3 is shown via "pip3 list", including tensorflow, pytorch, etc:
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
Package Version ---------------------------------- ------------------- absl-py 1.0.0 alabaster 0.7.12 anaconda-client 1.7.2 anaconda-navigator 2.0.3 anaconda-project 0.9.1 anyio 2.2.0 appdirs 1.4.4 argh 0.26.2 argon2-cffi 20.1.0 asn1crypto 1.4.0 astroid 2.5 astropy 4.2.1 astunparse 1.6.3 async-generator 1.10 atomicwrites 1.4.0 attrs 20.3.0 autopep8 1.5.6 Babel 2.9.0 backcall 0.2.0 backports.functools-lru-cache 1.6.4 backports.shutil-get-terminal-size 1.0.0 backports.tempfile 1.0 backports.weakref 1.0.post1 beautifulsoup4 4.9.3 bitarray 2.1.0 bkcharts 0.2 black 19.10b0 bleach 3.3.0 bokeh 2.3.2 boto 2.49.0 Bottleneck 1.3.2 brotlipy 0.7.0 cachetools 5.0.0 certifi 2020.12.5 cffi 1.14.5 chardet 4.0.0 click 8.1.2 cloudpickle 1.6.0 clyent 1.2.2 colorama 0.4.4 conda 4.10.1 conda-build 3.21.4 conda-content-trust 0+unknown conda-package-handling 1.7.3 conda-repo-cli 1.0.4 conda-token 0.3.0 conda-verify 3.4.2 contextlib2 0.6.0.post1 cryptography 3.4.7 cycler 0.10.0 Cython 0.29.23 cytoolz 0.11.0 dask 2021.4.0 decorator 5.0.6 defusedxml 0.7.1 diff-match-patch 20200713 distributed 2021.4.1 docutils 0.17.1 entrypoints 0.3 et-xmlfile 1.0.1 fastcache 1.1.0 filelock 3.0.12 flake8 3.9.0 Flask 1.1.2 flatbuffers 2.0 fsspec 0.9.0 future 0.18.2 gast 0.5.3 gevent 21.1.2 glob2 0.7 globus-cli 3.4.0 globus-sdk 3.5.0 gmpy2 2.0.8 google-auth 2.6.6 google-auth-oauthlib 0.4.6 google-pasta 0.2.0 greenlet 1.0.0 grpcio 1.46.0 h5py 2.10.0 HeapDict 1.0.1 html5lib 1.1 idna 2.10 imageio 2.9.0 imagesize 1.2.0 importlib-metadata 4.11.3 iniconfig 1.1.1 intervaltree 3.1.0 ipykernel 5.3.4 ipython 7.22.0 ipython-genutils 0.2.0 ipywidgets 7.6.3 isort 5.8.0 itsdangerous 1.1.0 jax 0.3.9 jdcal 1.4.1 jedi 0.17.2 jeepney 0.6.0 Jinja2 2.11.3 jmespath 0.10.0 joblib 1.0.1 json5 0.9.5 jsonschema 3.2.0 jupyter 1.0.0 jupyter-client 6.1.12 jupyter-console 6.4.0 jupyter-core 4.7.1 jupyter-packaging 0.7.12 jupyter-server 1.4.1 jupyterlab 3.0.14 jupyterlab-pygments 0.1.2 jupyterlab-server 2.4.0 jupyterlab-widgets 1.0.0 keras 2.8.0 Keras-Preprocessing 1.1.2 keyring 22.3.0 kiwisolver 1.3.1 lazy-object-proxy 1.6.0 libarchive-c 2.9 libclang 14.0.1 llvmlite 0.36.0 locket 0.2.1 lxml 4.6.3 Markdown 3.3.6 MarkupSafe 1.1.1 matplotlib 3.3.4 mccabe 0.6.1 mistune 0.8.4 mkl-fft 1.3.0 mkl-random 1.2.1 mkl-service 2.3.0 mock 4.0.3 more-itertools 8.7.0 mpmath 1.2.1 msgpack 1.0.2 multipledispatch 0.6.0 mypy-extensions 0.4.3 navigator-updater 0.2.1 nbclassic 0.2.6 nbclient 0.5.3 nbconvert 6.0.7 nbformat 5.1.3 nest-asyncio 1.5.1 networkx 2.5 nltk 3.6.1 nose 1.3.7 notebook 6.3.0 numba 0.53.1 numexpr 2.7.3 numpy 1.20.1 numpydoc 1.1.0 oauthlib 3.2.0 olefile 0.46 openpyxl 3.0.7 opt-einsum 3.3.0 packaging 20.9 pandas 1.2.4 pandocfilters 1.4.3 parso 0.7.0 partd 1.2.0 path 15.1.2 pathlib2 2.3.5 pathspec 0.7.0 patsy 0.5.1 pep8 1.7.1 pexpect 4.8.0 pickleshare 0.7.5 Pillow 8.2.0 pip 21.0.1 pkginfo 1.7.0 pluggy 0.13.1 ply 3.11 prometheus-client 0.10.1 prompt-toolkit 3.0.17 protobuf 3.20.1 psutil 5.8.0 ptyprocess 0.7.0 py 1.10.0 pyasn1 0.4.8 pyasn1-modules 0.2.8 pycodestyle 2.6.0 pycosat 0.6.3 pycparser 2.20 pycurl 7.43.0.6 pydocstyle 6.0.0 pyerfa 1.7.3 pyflakes 2.2.0 Pygments 2.8.1 PyJWT 2.3.0 pylint 2.7.4 pyls-black 0.4.6 pyls-spyder 0.3.2 pyodbc 4.0.0-unsupported pyOpenSSL 20.0.1 pyparsing 2.4.7 pyrsistent 0.17.3 PySocks 1.7.1 pytest 6.2.3 python-dateutil 2.8.1 python-jsonrpc-server 0.4.0 python-language-server 0.36.2 pytz 2021.1 PyWavelets 1.1.1 pyxdg 0.27 PyYAML 5.4.1 pyzmq 20.0.0 QDarkStyle 2.8.1 QtAwesome 1.0.2 qtconsole 5.0.3 QtPy 1.9.0 regex 2021.4.4 requests 2.25.1 requests-oauthlib 1.3.1 rope 0.18.0 rsa 4.8 Rtree 0.9.7 ruamel-yaml-conda 0.15.100 scikit-image 0.18.1 scikit-learn 0.24.1 scipy 1.6.2 seaborn 0.11.1 SecretStorage 3.3.1 Send2Trash 1.5.0 setuptools 52.0.0.post20210125 simplegeneric 0.8.1 singledispatch 0.0.0 sip 4.19.13 six 1.15.0 sniffio 1.2.0 snowballstemmer 2.1.0 sortedcollections 2.1.0 sortedcontainers 2.3.0 soupsieve 2.2.1 Sphinx 4.0.1 sphinxcontrib-applehelp 1.0.2 sphinxcontrib-devhelp 1.0.2 sphinxcontrib-htmlhelp 1.0.3 sphinxcontrib-jsmath 1.0.1 sphinxcontrib-qthelp 1.0.3 sphinxcontrib-serializinghtml 1.1.4 sphinxcontrib-websupport 1.2.4 spyder 4.2.5 spyder-kernels 1.10.2 SQLAlchemy 1.4.15 statsmodels 0.12.2 sympy 1.8 tables 3.6.1 tblib 1.7.0 tensorboard 2.8.0 tensorboard-data-server 0.6.1 tensorboard-plugin-wit 1.8.1 tensorflow 2.8.0 tensorflow-io-gcs-filesystem 0.25.0 termcolor 1.1.0 terminado 0.9.4 testpath 0.4.4 textdistance 4.2.1 tf-estimator-nightly 2.8.0.dev2021122109 threadpoolctl 2.1.0 three-merge 0.1.1 tifffile 2020.10.1 toml 0.10.2 toolz 0.11.1 torch 1.11.0 tornado 6.1 tqdm 4.59.0 traitlets 5.0.5 typed-ast 1.4.2 typing-extensions 4.1.1 ujson 4.0.2 unicodecsv 0.14.1 urllib3 1.26.4 watchdog 1.0.2 wcwidth 0.2.5 webencodings 0.5.1 Werkzeug 1.0.1 wheel 0.36.2 widgetsnbextension 3.5.1 wrapt 1.12.1 wurlitzer 2.1.0 xlrd 2.0.1 XlsxWriter 1.3.8 xlwt 1.3.0 xmltodict 0.12.0 yapf 0.31.0 zict 2.0.0 zipp 3.4.1 zope.event 4.5.0 zope.interface 5.3.0 |
Jupyter notebooks
The jupyter notebook executables are in your $PATH after loading the anaconda3 module. Don't run jupyter on the shared login nodes. Instead, follow these steps to attach a jupyter notebook running on a compute node to your local web browser:
1) Start a jupyter job via srun and note the hostname (you pick the port number for --port). |
Use the 2nd URL in step 3. Note the internal hostname in the cluster for step 2. | |||||
| 2) From your local desktop or laptop create an ssh tunnel to the compute node via a login node of delta. |
Authenticate with your login and 2-factor as usual. | |||||
3) Paste the 2nd URL (containing 127.0.0.1 and the token string) from step 1 into your browser and you will be connected to the jupyter instance running on your compute node of Delta.
|
|
Python (a recent or latest version)
If you do not need all of the extra modules provided by Anaconda, use the basic python installation under the gcc module. You can add modules via "pip3 install --user <modulename>", setup virtual environments, and customize as needed for your workflow but starting from a smaller installed base of python than Anaconda.
| Code Block | ||
|---|---|---|
| ||
$ module load gcc python $ which python /sw/spack/delta-2022-03/apps/python/3.10.4-gcc-11.2.0-3cjjp6w/bin/python $ module list Currently Loaded Modules: 1) modtree/gpu 3) gcc/11.2.0 5) ucx/1.11.2 7) python/3.10.4 2) default 4) cuda/11.6.1 6) openmpi/4.1.2 |
This is the list of modules available in the python from "pip3 list":
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
Package Version ------------------ --------- certifi 2021.10.8 cffi 1.15.0 charset-normalizer 2.0.12 click 8.1.2 cryptography 36.0.2 globus-cli 3.4.0 globus-sdk 3.5.0 idna 3.3 jmespath 0.10.0 pip 22.0.4 pycparser 2.21 PyJWT 2.3.0 requests 2.27.1 setuptools 58.1.0 urllib3 1.26.9 |
Launching Applications
- Launching One Serial Application
- Launching One Multi-Threaded Application
- Launching One MPI Application
- Launching One Hybrid (MPI+Threads) Application
- More Than One Serial Application in the Same Job
- MPI Applications One at a Time
- More than One MPI Application Running Concurrently
- More than One OpenMP Application Running Concurrently
Running Jobs
Job Accounting
The charge unit for Delta is the Service Unit (SU). This corresponds to the equivalent use of one compute core utilizing less than or equal to 2G of memory for one hour, or 1 GPU or fractional GPU using less than the corresponding amount of memory or cores for 1 hour (see table below). Keep in mind that your charges are based on the resources that are reserved for your job and don't necessarily reflect how the resources are used. Charges are based on either the number of cores or the fraction of the memory requested, whichever is larger. The minimum charge for any job is 1 SU.
Node Type | Service Unit Equivalence | |||
|---|---|---|---|---|
| Cores | GPU Fraction | Host Memory | ||
| CPU Node | 1 | N/A | 2 GB | |
GPU Node | Quad A100 | 2 | 1/7 A100 | 8 GB |
| Quad A40 | 16 | 1 A40 | 64 GB | |
| 8-way A100 | 2 | 1/7 A100 | 32 GB | |
| 8-way MI100 | 16 | 1 MI100 | 256 GB | |
Please note that a weighting factor will discount the charge for the reduced-precision A40 nodes, as well as the novel AMD MI100 based node - this will be documented through the XSEDE SU converter.
Local Account Charging
Use the accounts command to list the accounts available for charging. CPU and GPU resources will have individual charge names. For example in the following, abcd-delta-cpu and abcd-delta-gpu are available for user gbauer to use for the CPU and GPU resources.
| Code Block | ||
|---|---|---|
| ||
$ accounts available Slurm accounts for user gbauer: abcd-delta-cpu my_prefix my project abcd-delta-gpu my_prefix my project |
Job Accounting Considerations
- A node-exclusive job that runs on a compute node for one hour will be charged 128 SUs (128 cores x 1 hour)
- A node-exclusive job that runs on a 4-way GPU node for one hour will be charge 4 SUs (4 GPU x 1 hour)
- A node-exclusive job that runs on a 8-way GPU node for one hour will be charge 8 SUs (8 GPU x 1 hour)
- A shared job that runs on an A100 node will be charged for the fractional usage of the A100 (eg, using 1/7 of an A100 for one hour will be 1/7 GPU x 1 hour, or 1/7 SU per hour, except the first hour will be 1 SU (minimum job charge).
Accessing the Compute Nodes
Delta implements the Slurm batch environment to manage access to the compute nodes. Use the Slurm commands to run batch jobs or for interactive access to compute nodes. See: https://slurm.schedmd.com/quickstart.html for an introduction to Slurm. There are two ways to access compute nodes on Delta.
Batch jobs can be used to access compute nodes. Slurm provides a convenient direct way to submit batch jobs. See https://slurm.schedmd.com/heterogeneous_jobs.html#submitting for details.
Sample Slurm batch job scripts are provided in the Job Scripts section below.
Direct ssh access to a compute node in a running batch job from a dt-loginNN node is enabled, once the job has started.
| Code Block |
|---|
$ squeue --job jobid
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
12345 cpu bash gbauer R 0:17 1 cn001 |
Then in a terminal session:
| Code Block |
|---|
$ ssh cn001 cn001.delta.internal.ncsa.edu (172.28.22.64) OS: RedHat 8.4 HW: HPE CPU: 128x RAM: 252 GB Site: mgmt Role: compute $ |
Scheduler
For information, consult:
https://slurm.schedmd.com/quickstart.html
slurm quick reference guide View file name slurm_summary.pdf height 250
Partitions (Queues)
Table. Delta Production Partitions/Queues
Partition/Queue | Node Type | Max Nodes per Job | Max Duration | Max Jobs in Queue* | Charge Factor |
|---|---|---|---|---|---|
cpu | CPU | TBD | 24 hr / 48 hr | TDB | 1.0 |
| cpu-interactive | CPU | TBD | 30 min | TBD | 2.0 |
| gpuA100x4 | quad A100 | TBD | 24 hr / 48 hr | TDB | 1.0 |
| gpuA100x4-interactive | quad-A100 | TBD | 30 min | TBD | 2.0 |
| gpuA100x8 | octa-A100 | TBD | 24 hr / 48 hr | TDB | 1.0 |
| gpuA100x8-interactive | octa-A100 | TBD | 30 min | TBD | 2.0 |
| gpuA40x4 | quad-A40 | TBD | 24 hr / 48 hr | TBD | 0.6 |
| gpuA40x4-interactive | quad-A40 | TBD | 30 min | TBD | 1.2 |
| gpuMI100x8 | octa-MI100 | TBD | 24 hr / 48 hr | TBD | 1.0 |
| gpuMI100x8-interactive | octa-MI100 | TBD | 30 min | TBD | 2.0 |
sview view of slurm partitions
Node Policies
Node-sharing is the default for jobs. Node-exclusive mode can be obtained by specifying all the consumable resources for that node type or adding the following Slurm options:
--exclusive --mem=0
GPU NVIDIA MIG (GPU slicing) for the A100 will be supported at a future date.
Pre-emptive jobs will be supported at a future date.
Interactive Sessions
Interactive sessions can be implemented in several ways depending on what is needed.
To start up a bash shell terminal on a cpu or gpu node
- single core with 1GB of memory, with one task on a cpu node
| Code Block |
|---|
srun --account=account_name --partition=cpu \ --nodes=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=1g \ --pty bash |
- single core with 20GB of memory, with one task on a A40 gpu node
| Code Block |
|---|
srun --account=account_name --partition=gpuA40x4 \ --nodes=1 --gpus-per-node=1 --tasks=1 \ --tasks-per-node=1 --cpus-per-task=1 --mem=20g \ --pty bash |
| Info | ||
|---|---|---|
| ||
Since interactive jobs are already a child process of srun, one cannot srun applications from within them. Use mpirun to launch mpi jobs from within an interactive job. Within standard batch jobs submitted via sbatch, use srun to launch MPI codes. |
Interactive X11 Support
To run an X11 based application on a compute node in an interactive session, the use of the --x11 switch with srun is needed. For example, to run a single core job that uses 1g of memory with X11 (in this case an xterm) do the following:
| Code Block |
|---|
srun -A abcd-delta-cpu --partition=cpu \ --nodes=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=1g \ --x11 xterm |
Sample Job Scripts Anchor Job Scripts Job Scripts
| Job Scripts | |
| Job Scripts |
Serial jobs
Code Block language bash title serial example script $ cat job.slurm #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=1 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=myjobtest #SBATCH --time=00:10:00 # hh:mm:ss for the job ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module purge # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) module load python # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" srun python3 myprog.pyMPI
Code Block language bash title mpi example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=2 #SBATCH --ntasks-per-node=32 #SBATCH --cpus-per-task=1 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=mympi #SBATCH --time=00:10:00 # hh:mm:ss for the job ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module purge # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) module load gcc/11.2.0 openmpi # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" srun osu_reduceOpenMP
Code Block language bash title openmp example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=32 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=myopenmp #SBATCH --time=00:10:00 # hh:mm:ss for the job ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module purge # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) module load gcc/11.2.0 # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" export OMP_NUM_THREADS=32 srun stream_gccHybrid (MPI + OpenMP or MPI+X)
Code Block language bash title mpi+x example script collapse true #!/bin/bash #SBATCH --mem=16g #SBATCH --nodes=2 #SBATCH --ntasks-per-node=4 #SBATCH --cpus-per-task=4 # <- match to OMP_NUM_THREADS #SBATCH --partition=cpu # <- or one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 #SBATCH --account=account_name #SBATCH --job-name=mympi+x #SBATCH --time=00:10:00 # hh:mm:ss for the job ### GPU options ### ##SBATCH --gpus-per-node=2 ##SBATCH --gpu-bind=none # <- or closest ##SBATCH --mail-user=you@yourinstitution.edu ##SBATCH --mail-type="BEGIN,END" See sbatch or srun man pages for more email options module purge # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) module load gcc/11.2.0 openmpi # ... or any appropriate modules module list # job documentation and metadata echo "job is starting on `hostname`" export OMP_NUM_THREADS=4 srun xthi- Parametric / Array / HTC jobs
Job Management
Batch jobs are submitted through a job script (as in the examples above) using the sbatch command. Job scripts generally start with a series of SLURM directives that describe requirements of the job such as number of nodes, wall time required, etc… to the batch system/scheduler (SLURM directives can also be specified as options on the sbatch command line; command line options take precedence over those in the script). The rest of the batch script consists of user commands.
The syntax for sbatch is:
sbatch [list of sbatch options] script_name
Refer to the sbatch man page for detailed information on the options.
squeue/scontrol/sinfo
Commands that display batch job and partition information .
| SLURM EXAMPLE COMMAND | DESCRIPTION |
|---|---|
| squeue -a | List the status of all jobs on the system. |
| squeue -u $USER | List the status of all your jobs in the batch system. |
| squeue -j JobID | List nodes allocated to a running job in addition to basic information.. |
| scontrol show job JobID | List detailed information on a particular job. |
| sinfo -a | List summary information on all the partition. |
See the manual (man) pages for other available options.
Useful Batch Job Environment Variables
| Anchor | ||||
|---|---|---|---|---|
|
DESCRIPTION | SLURM ENVIRONMENT VARIABLE | DETAIL DESCRIPTION |
|---|---|---|
| JobID | $SLURM_JOB_ID | Job identifier assigned to the job |
| Job Submission Directory | $SLURM_SUBMIT_DIR | By default, jobs start in the directory that the job was submitted from. So the "cd $SLURM_SUBMIT_DIR" command is not needed. |
| Machine(node) list | $SLURM_NODELIST | variable name that contains the list of nodes assigned to the batch job |
| Array JobID | $SLURM_ARRAY_JOB_ID $SLURM_ARRAY_TASK_ID | each member of a job array is assigned a unique identifier |
See the sbatch man page for additional environment variables available.
srun
The srun command initiates an interactive job on the compute nodes.
For example, the following command:
srun -A account_name --time=00:30:00 --nodes=1 --ntasks-per-node=64 --pty /bin/bash
will run an interactive job in the default queue with a wall clock limit of 30 minutes, using one node and 16 cores per node. You can also use other sbatch options such as those documented above.
After you enter the command, you will have to wait for SLURM to start the job. As with any job, your interactive job will wait in the queue until the specified number of nodes is available. If you specify a small number of nodes for smaller amounts of time, the wait should be shorter because your job will backfill among larger jobs. You will see something like this:
srun: job 123456 queued and waiting for resources
Once the job starts, you will see:
srun: job 123456 has been allocated resources
and will be presented with an interactive shell prompt on the launch node. At this point, you can use the appropriate command to start your program.
When you are done with your work, you can use the exit command to end the job.
scancel
The scancel command deletes a queued job or terminates a running job.
- scancel JobID deletes/terminates a job.
Refunds
Refunds are considered, when appropriate, for jobs that failed due to circumstances beyond user control.
XSEDE users and project that wish to request a refund should see the XSEDE Refund Policy section located here.
Other allocated users and projects wishing to request a refund should email help@ncsa.illinois.edu. Please include the batch job ids and the standard error and output files produced by the job(s).
Visualization
Delta A40 nodes support NVIDIA raytracing hardware.
- describe visualization capabilities & software.
- how to establish VNC/DVC/remote desktop
Containers
Singularity
Container support on Delta is provided by Singularity.
Docker images can be converted to Singularity sif format via the singularity pull command. Commands can be run from within a container using singularity run command.
If you encounter quota issues with Singularity caching in ~/.singularity , the environment variable SINGULARITY_CACHEDIR can be used to use a different location such as a scratch space.
NVIDIA NGC Containers
Delta provides NVIDIA NGC Docker containers that we have pre-built with Singularity. Look for the latest binary containers in /sw/external/NGC/ . The containers are used as shown in the sample scripts below:
| Code Block | ||||
|---|---|---|---|---|
| ||||
#!/bin/bash #SBATCH --mem=16g64g #SBATCH --nodes=1 #SBATCH --ntasks-per-node=1 #SBATCH --cpus-per-task=164 # <- match to OMP_NUM_THREADS, 64 requests whole node #SBATCH --partition=gpuA100x4 # <- one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8 ##SBATCH#SBATCH --account=account_namebbka-delta-gpu #SBATCH --job-name=pytorchNGC ### GPU options ### #SBATCH --gpus-per-node=1 #SBATCH --gpus-per-task=1 #SBATCH --gpu-bind=none # <- or closestverbose,per_task:1 module purge # drop modules and explicitly load the ones needed # (good job metadata and reproducibility) module list # job documentation and metadata echo "job is starting on `hostname`" # run the container binary with arguments: python3 <program.py> singularity run --nv \ /sw/external/NGC/pytorch:22.02-py3 python3 tensor_gpu.py |
| Code Block | ||||||
|---|---|---|---|---|---|---|
| ||||||
#!/bin/bash
#SBATCH --mem=16g
#SBATCH --nodes=1
#SBATCH --ntasks-per-node=1
#SBATCH --cpus-per-task=1 # <- match to OMP_NUM_THREADS
#SBATCH --partition=gpuA100x4 # <- one of: gpuA100x4 gpuA40x4 gpuA100x8 gpuMI100x8
##SBATCH --account=account_name
#SBATCH --job-name=tfNGC
### GPU options ###
#SBATCH --gpus-per-node=2
#SBATCH --gpu-bind=none # <- or closest
module purge # drop modules and explicitly load the ones needed
# (good job metadata and reproducibility)
module list # job documentation and metadata
echo "job is starting on `hostname`"
# run the container binary with arguments: python3 <program.py>
/sw/external/NGC/tensorflow:22.02-tf2-py3 python3 tf_gpu.py |
Container list (as of March, 2022)
| Code Block | ||
|---|---|---|
| ||
caffe:20.03-py3 caffe2:18.08-py3 cntk:18.08-py3 , Microsoft Cognitive Toolkit digits:21.09-tensorflow-py3 matlab:r2021b mxnet:21.09-py3 pytorch:22.02-py3 tensorflow:22.02-tf1-py3 tensorflow:22.02-tf2-py3 tensorrt:22.02-py3 theano:18.08 torch:18.08-py2 |
see also: https://catalog.ngc.nvidia.com/orgs/nvidia/containers
Other Containers
Extreme-scale Scientific Software Stack (E4S)
The E4S container with GPU (cuda and rocm) support is provided for users of specific ECP packages made available by the E4S project (https://e4s-project.github.io/). The singularity image is available as :
/sw/external/E4S/e4s-gpu-x86_64.sifTo use E4S with NVIDIA GPUs
| Code Block |
|---|
$ srun --account=account_name --partition=gpuA100-interactive \ --nodes=1 --gpus-per-node=1 --tasks=1 --tasks-per-node=1 \ --cpus-per-task=1 --mem=20g \ --pty bash $ singularity exec --cleanenv /sw/external/E4S/e4s-gpu-x86_64.sif \ /bin/bash --rcfile /etc/bash.bashrc |
The spack package inside of the image will interact with a local spack installation. If ~/.spack directory exists, it might need to be renamed.
More information can be found at https://e4s-project.github.io/download.html
Protected Data (N/A)
...
Help
For assistance with the use of Delta
- XSEDE users can create a ticket via the user portal at https://portal.xsede.org/web/xup/help-desk
- All other users (Illinois allocations, Diversity Allocations, etc) please send email to help@ncsa.illinois.edu.
Acknowledge
To acknowledge the NCSA Delta system in particular, please include the following
This research is part of the Delta research computing project, which is supported by the National Science Foundation (award OCI 2005572), and the State of Illinois. Delta is a joint effort of the University of Illinois at Urbana-Champaign and its National Center for Supercomputing Applications.
To include acknowledgement of XSEDE contributions to a publication or presentation please see https://portal.xsede.org/acknowledge and https://www.xsede.org/for-users/acknowledgement.
References
Supporting documentation resources:
https://www.rcac.purdue.edu/knowledge/anvil
https://nero-docs.stanford.edu/jupyter-slurm.html